COX-2 Expression in Breast Cancer and Impact on Survival Outcomes
Article Information
Daniely Regina Freitas-Alves1, Juliana Batoca Pinto2, Fabiana Resende Rodrigues3, Priscila Valverde3, Danielle Clemente da Silva Fernandes3, Maria Thereza Accioly4, Samuel dos Santos Valença2, Jamila Alessandra Perini1,5, Rosane Vianna-Jorge1,2*
1Programa de Saúde Pública e Meio Ambiente - Escola Nacional de Saúde Pública - FIOCRUZ, Rio de Janeiro, RJ, Brasil
2Instituto de Ciências Biomédicas, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil
3Divisão de Patologia, Instituto Nacional do Câncer, Rio de Janeiro, RJ, Brasil
4Banco Nacional de Tumores, Instituto Nacional do Câncer, Rio de Janeiro, RJ, Brasil
5Laboratório de Pesquisa de Ciências Farmacêuticas, Unidade de Farmácia, Centro Universitário Estadual da Zona Oeste, Rio de Janeiro, RJ, Brasil
*Corresponding Author: Prof. Rosane Vianna-Jorge, Programa de Farmacologia e Inflamação, Instituto de Ciências Biomédicas, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brasil-Av. Carlos Chagas Filho, 373, Bloco J, sala 27, Centro de Ciências da Saúde, Cidade Universitária, Ilha do Fundão, Rio de Janeiro, RJ, Brasil
Received: 27 April 2020; Accepted: 11 May 2020; Published: 20 May 2020
Citation: Daniely Regina Freitas-Alves, Juliana Batoca Pinto, Fabiana Resende Rodrigues, Priscila Valverde, Danielle Clemente da Silva Fernandes, Maria Thereza Accioly, Samuel dos Santos Valença, Jamila Alessandra Perini, Rosane Vianna-Jorge. COX-2 Expression in Breast Cancer and Impact on Survival Outcomes. Journal of Cancer Science and Clinical Therapeutics 4 (2020): 120-132.
View / Download Pdf Share at FacebookAbstract
Purpose: The inducible inflammatory enzyme cyclooxygenase-2 (COX-2) favors carcinogenesis, but its expression in breast tumors presents great variability, with controversial prognostic impact. Here, we characterize COX-2 protein levels in breast tumors by immunohistochemistry according to gene polymorphisms, and evaluate if tumor COX-2 protein levels or mRNA are associated with survival outcomes.
Methods: First, COX-2 protein levels were quantified by immunohistochemistry in selected tissue specimens (N=236) of excised breasts from a hospital-based cohort of breast cancer in Brazil, and evaluated for their association with gene polymorphisms and histopathological variables, as well as with survival outcomes. Secondly, an online gene array database compiling information from different breast cancer cohorts was used to analyze the association between tumor COX-2 mRNA and survival outcomes.
Results: High COX-2 protein levels were associated with high tumor grade (OR=1.86; 95% CI=1.1-3.17), but not with gene variants or survival outcomes. In contrast, high COX-2 mRNA was associated with better disease-free survival when considering all cases (HR=0.82; 95%CI=0.72-0.92; N=3951) or only ER+ tumors (HR=0.62; 95%CI=0.49-0.79; N=2061), but with worse disease-free survival (HR=1.6; 95%CI=1.22-2.11; N=618) among patients with basal-like tumors.
Conclusion: Gene polymorphisms do not account for the variability on COX-2 protein levels in breast tumors, and COX-2 mRNA seems to be a better candidate for prognostic evaluation of breast cancer survival, but its impact depends on breast cancer subtypes.
Keywords
COX-2; Breast cancer survival; PTGS2; Gene polymorphisms
COX-2 articles, Breast cancer survival articles, PTGS2 articles, Gene polymorphisms articles
COX-2 articles COX-2 Research articles COX-2 review articles COX-2 PubMed articles COX-2 PubMed Central articles COX-2 2023 articles COX-2 2024 articles COX-2 Scopus articles COX-2 impact factor journals COX-2 Scopus journals COX-2 PubMed journals COX-2 medical journals COX-2 free journals COX-2 best journals COX-2 top journals COX-2 free medical journals COX-2 famous journals COX-2 Google Scholar indexed journals Breast cancer survival articles Breast cancer survival Research articles Breast cancer survival review articles Breast cancer survival PubMed articles Breast cancer survival PubMed Central articles Breast cancer survival 2023 articles Breast cancer survival 2024 articles Breast cancer survival Scopus articles Breast cancer survival impact factor journals Breast cancer survival Scopus journals Breast cancer survival PubMed journals Breast cancer survival medical journals Breast cancer survival free journals Breast cancer survival best journals Breast cancer survival top journals Breast cancer survival free medical journals Breast cancer survival famous journals Breast cancer survival Google Scholar indexed journals PTGS2 articles PTGS2 Research articles PTGS2 review articles PTGS2 PubMed articles PTGS2 PubMed Central articles PTGS2 2023 articles PTGS2 2024 articles PTGS2 Scopus articles PTGS2 impact factor journals PTGS2 Scopus journals PTGS2 PubMed journals PTGS2 medical journals PTGS2 free journals PTGS2 best journals PTGS2 top journals PTGS2 free medical journals PTGS2 famous journals PTGS2 Google Scholar indexed journals Gene polymorphisms articles Gene polymorphisms Research articles Gene polymorphisms review articles Gene polymorphisms PubMed articles Gene polymorphisms PubMed Central articles Gene polymorphisms 2023 articles Gene polymorphisms 2024 articles Gene polymorphisms Scopus articles Gene polymorphisms impact factor journals Gene polymorphisms Scopus journals Gene polymorphisms PubMed journals Gene polymorphisms medical journals Gene polymorphisms free journals Gene polymorphisms best journals Gene polymorphisms top journals Gene polymorphisms free medical journals Gene polymorphisms famous journals Gene polymorphisms Google Scholar indexed journals prostaglandin E2 articles prostaglandin E2 Research articles prostaglandin E2 review articles prostaglandin E2 PubMed articles prostaglandin E2 PubMed Central articles prostaglandin E2 2023 articles prostaglandin E2 2024 articles prostaglandin E2 Scopus articles prostaglandin E2 impact factor journals prostaglandin E2 Scopus journals prostaglandin E2 PubMed journals prostaglandin E2 medical journals prostaglandin E2 free journals prostaglandin E2 best journals prostaglandin E2 top journals prostaglandin E2 free medical journals prostaglandin E2 famous journals prostaglandin E2 Google Scholar indexed journals carcinogenesis articles carcinogenesis Research articles carcinogenesis review articles carcinogenesis PubMed articles carcinogenesis PubMed Central articles carcinogenesis 2023 articles carcinogenesis 2024 articles carcinogenesis Scopus articles carcinogenesis impact factor journals carcinogenesis Scopus journals carcinogenesis PubMed journals carcinogenesis medical journals carcinogenesis free journals carcinogenesis best journals carcinogenesis top journals carcinogenesis free medical journals carcinogenesis famous journals carcinogenesis Google Scholar indexed journals immunohistochemistry articles immunohistochemistry Research articles immunohistochemistry review articles immunohistochemistry PubMed articles immunohistochemistry PubMed Central articles immunohistochemistry 2023 articles immunohistochemistry 2024 articles immunohistochemistry Scopus articles immunohistochemistry impact factor journals immunohistochemistry Scopus journals immunohistochemistry PubMed journals immunohistochemistry medical journals immunohistochemistry free journals immunohistochemistry best journals immunohistochemistry top journals immunohistochemistry free medical journals immunohistochemistry famous journals immunohistochemistry Google Scholar indexed journals polymorphisms articles polymorphisms Research articles polymorphisms review articles polymorphisms PubMed articles polymorphisms PubMed Central articles polymorphisms 2023 articles polymorphisms 2024 articles polymorphisms Scopus articles polymorphisms impact factor journals polymorphisms Scopus journals polymorphisms PubMed journals polymorphisms medical journals polymorphisms free journals polymorphisms best journals polymorphisms top journals polymorphisms free medical journals polymorphisms famous journals polymorphisms Google Scholar indexed journals nucleotide polymorphisms articles nucleotide polymorphisms Research articles nucleotide polymorphisms review articles nucleotide polymorphisms PubMed articles nucleotide polymorphisms PubMed Central articles nucleotide polymorphisms 2023 articles nucleotide polymorphisms 2024 articles nucleotide polymorphisms Scopus articles nucleotide polymorphisms impact factor journals nucleotide polymorphisms Scopus journals nucleotide polymorphisms PubMed journals nucleotide polymorphisms medical journals nucleotide polymorphisms free journals nucleotide polymorphisms best journals nucleotide polymorphisms top journals nucleotide polymorphisms free medical journals nucleotide polymorphisms famous journals nucleotide polymorphisms Google Scholar indexed journals solid tumors articles solid tumors Research articles solid tumors review articles solid tumors PubMed articles solid tumors PubMed Central articles solid tumors 2023 articles solid tumors 2024 articles solid tumors Scopus articles solid tumors impact factor journals solid tumors Scopus journals solid tumors PubMed journals solid tumors medical journals solid tumors free journals solid tumors best journals solid tumors top journals solid tumors free medical journals solid tumors famous journals solid tumors Google Scholar indexed journals
Article Details
Abbrevations:
COX-2- cyclooxygenase-2; PGE2- prostaglandin E2; SNPs- single nucleotide polymorphisms; INCA-Brazil- Brazilian National Cancer Institute; CS- continuous scale; IRS- immunoreaction score; OR- odds ratio; HR- hazard ratio; ER- Estrogen Receptor; HER2- Human Epidermal Growth Factor Receptor 2
1. Background
Breast cancer is the most incident and prevalent cancer among women worldwide [1], and a highly heterogeneous disease, with diverse morphological and molecular presentations [2]. Although the advances in tumor classification and personalized treatment have contributed to reduce its global mortality [3], breast cancer remains the first cause of death by cancer among women [4]. As an attempt to identify additional molecular targets that may guide clinical conducts or improve prognostic evaluation, vital biological processes in breast carcinogenesis are under scrutiny [5].
Chronic inflammation is a hallmark of several cancers, since it ultimately contributes for tumor growth, migration and metastasis [6]. In breast cancer, the presence of an inflammatory infiltrate was first proposed as a prognostic marker by [7]. Since then, many inflammatory factors, as well as their receptors, have been shown to participate in various steps of tumor development, including cell proliferation, differentiation, angiogenesis and metastasis [8]. The inducible enzyme cyclooxygenase-2 (COX-2), which is coded by the PTGS2 gene, is recognized as the master switch that activates the inflammatory response; its induction leads to the biosynthesis of prostaglandins, particularly prostaglandin E2 (PGE2), which orchestrates the inflammatory response [9]. In invasive breast carcinoma, the frequencies of COX-2 overexpression range from 17% to 84% [10], and the mechanisms underlying this variability are not yet fully understood.
PTGS2 gene is highly regulated, both in the promoter [11] and in the 3′-untranslated [12] regions. PTGS2 is also highly polymorphic, with several single nucleotide polymorphisms (SNPs) in these regulatory regions [13-15]. The four most common PTGS2 SNPs (rs689465, rs689466, rs20417, and rs5275) have estimated global frequencies > 0.1 [16], and two of them (rs689466 and rs5275) have been shown to affect gene expression in in vitro studies. Thus, rs689466 (-1195 G variant) increased gene transcription in different cell models [17-19], whereas rs5275 (8473 C variant) appears to favor mRNA stability [13]. However, there are no in vivo studies evaluating the impact of these SNPs on tumor levels of COX-2, either in breast cancer or in other solid tumors.
A recent paper from our group suggests an association of rs689466 (-1195 G variant) with a significant reduction in disease-free survival of obese breast cancer patients [20]. The link between excess weight or obesity and breast cancer appears to involve altered expression of hormones, especially estrogen, as well as growth factors and inflammatory mediators, including PGE2 [21]. These findings favor the idea that chronic inflammation and induction of COX-2 in tumor microenvironment may contribute for worse prognosis of breast cancer.
Here, we evaluate if rs689466 and other major PTGS2 SNPs affect COX-2 protein levels in breast tumors, and if tumor COX-2, either as mRNA or protein levels, may contribute as a prognostic predictor of disease-free and overall survival of breast cancer patients. Two approaches were used. First, COX-2 was quantified by immunohistochemistry in selected tissue specimens of excised breasts from a hospital-based cohort of breast cancer in Brazil, whose patients had been previously genotyped for PTGS2. COX-2 protein levels were evaluated for their association with PTGS2 SNPs and histopathological variables, as well as for their impact on disease-free and overall survival. Second, an online gene array database (compiling GEO, EGA and TCGA platforms) was assessed via the online software KMplotter (www.kmplot.com) [22] to analyze the association between tumor COX-2 mRNA and survival outcomes of different breast cancer cohorts.
2. Materials and Methods
2.1 COX-2 evaluation in breast tumors from a single hospital-based cohort
2.1.1 Tumor selection: Tumor blocks (N=236) were selected from a hospital-based cohort of Brazilian women with first diagnosis of unilateral breast carcinoma and no distant metastases (N=713) who were assigned for curative surgery as their first therapeutic approach at the Brazilian National Cancer Institute (INCA-Brazil), during the period from February 2009 to April 2013. The study protocol was approved by the Ethics Committees of the Brazilian National Cancer Institute (INCA #129/08) and of the National School of Public Health (FIOCRUZ/CAAE 55929416.8.0000.5240), and all patients gave written consent to participate. The description of this cohort formation and its main clinical characteristics have been previously published [23, 24]. All patients were genotyped for rs689465 (-1290AG), rs689466 (-1195AG), rs20417 (-765GC) and rs5275 (8473TC) [20].
The selection of tumor blocks was based on PTGS2 genotypes and breast cancer subtypes. The 236 tumors that were included comprising all available cases with variant rs689466 genotypes (-1195 AG + GG, N=114) and 123 tumor blocks from patients with the wild-type genotype (-1195 AA). All available blocks of HER2-like or Triple-Negative tumors (N=72) were also included (55 AA, 17 AG + GG).
2.1.2 Immunohistochemistry and scoring: Paraffin-embedded tumor samples were cut into sections of 3μm thick tissue and mounted on glass slides with 3-aminopropyl triethoxysilane (Sigma ChemicalCo, St. Louis, MO USA). The slides were deparaffinized in xylene baths at 25°C and rehydrated in a grading system of ethanol and water. Haematoxylin and eosin staining were performed to select the most representative specimen for each patient [25]. COX-2 detection was performed with monoclonal mouse anti-human COX-2 antibody, clone CX 294 (dilution 1:100) (Agilent Technologies Inc, Santa Clara, USA). Incubations were carried out overnight and then revealed using Novolink Polymer Detection System standard protocol (Leica Biosystems, Newcastle Ltd, USA). Colon adenocarcinoma was used for negative and positive controls. Negative controls were verified in the absence of the monoclonal antibody. Because of the observed intratumoral variability in COX-2 staining in breast tumors, the individual quantification included the whole area of a representative tumor slide and was based on the two previously published scoring methods: a continuous scale (CS) [26] and a categorical score, [25] both of which consider the percentage of immunostained cells and the intensity of the reaction.
The CS was calculated as follows: CS=(% weak × 1) + (% moderate × 2) + (% strong × 3) [26]. The intensity of COX-2 staining was rated as follows: negative (complete absence of cellular reaction); weak (diffuse and mild reaction in cytoplasm, with no detectable reaction in cell membranes); moderate (detectable reaction in both cytoplasm and plasma membrane); or strong (strong in both cytoplasm and plasma membrane). The categorical score, or immunoreaction score (IRS), was defined by the following equation: IRS=(positivity score) × (intensity score). The positivity score was attributed 1 to 4, according to the percentage of positive cancer cells: 1 (1%-9%), 2 (10%-49%), 3 (50%-79%), or 4 (80%-100%). The intensity score ranged 0-3: negative (0), weak (1), moderate (2), or strong (3) [25]. Breast tissues were considered positive for COX-2 when the IRS was ≥ 6, meaning that at least 10% of the cells presented moderate staining.
All slides were blindly evaluated by a pathologist (FRR), using a light microscope (Nikon, Tokyo, Japan). High quality images were captured using the Aperio ImageScope (Leica Biosystems, Newcastle Ltd, USA).
2.1.3 Survival outcomes: Disease relapse was defined as the primary clinical endpoint of the study, and was characterized by the occurrence of loco-regional or contra-lateral recurrence of breast cancer or by any distant metastasis. Disease-free survival was defined as the period of time between the date of surgery and the date of first relapse detection. Patients were considered disease-free if they had no suggestive clinical symptoms or imaging diagnosis of disease progression until their last medical consult. New primary cancer lesions were censored in the analysis of disease-free survival. Overall survival was the secondary clinical endpoint, and was considered as the period of time between the date of surgery and the date of death by any cause. Patients achieving five years of follow-up were censored for both disease-free and overall survival.
2.1.4 Statistical analyses: Histopathological variables and PTGS2 genotypes were categorized and expressed in numbers and relative frequencies. COX-2 expression based on CS was compared between categories of histopathological variables and PTGS2 genotypes using the Mann–Whitney U test. The association between IRS and PTGS2 genotypes or histopathological variables was evaluated with the x2 test, with the calculation of the odds ratios (ORs) and respective 95% CI. All the statistical analyses were conducted by using IBM SPSS Version 20 for Windows (IBM Corp., Armonk, NY, USA). The impact of individual variables on disease-free survival rates was estimated by calculation of their Hazard Ratios (HR), and 95% confidence intervals (95% CI), using Cox regression models. Descriptive statistics and survival analyses were conducted using SPSS 13.0 for Windows (SPSS Inc., Chicago, Illinois).
2.2 COX-2 mRNA and impact on survival outcomes from compiled breast cancer cohortsPublically available information regarding gene expression profiles, clinical data and survival outcomes of different breast cancer cohorts that are compiled in the GEO, EGA and TCGA platforms were assessed via the online software KMplotter (www.kmplot.com) [22]. Relapse-free survival and overall survival were analyzed using the filters for breast cancer and for PTGS2 gene, censoring the follow-up time in 60 months, and setting the best cut-off value of mRNA expression to categorize tumor expression as “low” or “high”. Additional filters were used to evaluate the results according to breast cancer subsets, as follows: Estrogen Receptor (ER) positive or negative; Human Epidermal Growth Factor Receptor 2 (HER2) positive and Triple Negative (negative for ER, HER2 and for the Progesterone Receptor-PR).
3. Results
COX-2 immunostaining was characterized in all selected tumors (N=236) from the INCA-Brazil cohort, and a representative image is shown in Figure 1. Figures 1a and 1b illustrate fully negative reactions, whereas Figures 1c to 1h show a gradation of the immunostaining intensity, characterized as weak (Figures 1c and 1d), moderate (Figures 1e and 1f), or strong (Figures 1g and 1h). The distribution of the CS showed a median of 100 with an interquartile range of 40-200 and, according to the IRS values, 129 (55%) breast tumors were considered with high COX-2 expression (IRS ≥ 6).
The distribution of CS and IRS was evaluated according to individual clinical features and to PTGS2 genotypes (Table 1). No significant associations were found, except for a higher proportion of positive COX-2 expression among high-grade (G3) tumors, although the median values and the distribution of CS were not different according to tumor grades.
|
Individual features |
IRS < 6 |
IRS ≥ 6 |
CS |
||||||
|
N* |
% |
N* |
% |
OR |
95%CI |
Median |
IR |
pM-W |
|
|
Grade |
|||||||||
|
G1 + G2 |
54 |
50.9 |
44 |
35.8 |
150 |
55.0 - 217.5 |
|||
|
G3 |
52 |
49.1 |
79 |
64.2 |
1.86 |
1.1 - 3.17 |
140 |
50.0 - 200.0 |
0.487 |
|
Tumor size |
|||||||||
|
pT1 |
47 |
43.9 |
55 |
43 |
100 |
50.0 - 218.75 |
|||
|
pT2 + pT3 |
60 |
56.1 |
73 |
57 |
1.04 |
0.62 - 1.75 |
150 |
50.0 - 200.0 |
0.866 |
|
Lymph node status |
|||||||||
|
pN0 + pN1 |
57 |
53.3 |
70 |
54.7 |
150 |
50.0 - 225.0 |
|||
|
pN2 + pN3 |
50 |
46.7 |
58 |
45.3 |
0.7 |
0.42 - 1.17 |
100 |
55.0 - 200.0 |
0.258 |
|
Stage |
|||||||||
|
I + II |
67 |
62.6 |
83 |
65.4 |
120 |
50.0 - 225.0 |
|||
|
III |
40 |
37.4 |
44 |
34.6 |
0.89 |
0.52 - 1.52 |
150 |
70.0 - 200.0 |
0.918 |
|
ER/PR |
|||||||||
|
Negative |
32 |
29.9 |
40 |
31 |
150 |
50.0 - 225.0 |
|||
|
Positive |
75 |
70.1 |
89 |
69 |
1.05 |
0.6 - 1.84 |
100 |
52.5 - 175.0 |
0.073 |
|
Biological classification |
|||||||||
|
Luminal |
75 |
70.1 |
89 |
69 |
150 |
50.0 - 225.0 |
|||
|
HER-2 + Triple negative |
32 |
29.9 |
40 |
31 |
1.05 |
0.6 - 1.84 |
100 |
52.5 - 175.0 |
0.073 |
|
Obesity |
|||||||||
|
Normal + Overweight |
77 |
72 |
91 |
70.5 |
140 |
50.0 - 200.0 |
|||
|
Obese |
30 |
28 |
38 |
29.5 |
1.07 |
0.61 - 1.89 |
140 |
60.0 - 200.0 |
0.952 |
|
Menopausal status |
|||||||||
|
Pre-menopausal |
29 |
27.4 |
29 |
22.7 |
100 |
50.0 - 200.0 |
|||
|
Post-menopausal |
77 |
72.6 |
99 |
77.3 |
1.29 |
0.71 - 2.33 |
140 |
50.0 - 200.0 |
0.461 |
|
PTGS2 genotypes |
|||||||||
|
1290AG |
|||||||||
|
AA |
91 |
85.1 |
107 |
82.9 |
100 |
47.5 - 200.0 |
|||
|
AG + GG |
16 |
14.9 |
22 |
17.1 |
1.17 |
0.58 - 2.36 |
160 |
100 - 213.75 |
0.146 |
|
1195AG |
|||||||||
|
AA |
51 |
47.7 |
72 |
55.8 |
100 |
70.0 - 200.0 |
|||
|
AG + GG |
56 |
52.3 |
57 |
44.2 |
0.72 |
0.43 - 1.21 |
160 |
37.5 - 225.0 |
0.655 |
|
765GC |
|||||||||
|
GG |
84 |
78.5 |
95 |
73.6 |
100 |
50 - 200.0 |
|||
|
GC + CC |
23 |
21.5 |
34 |
26.4 |
1.31 |
0.71 - 2.39 |
160 |
72.5 - 221.25 |
0.324 |
|
8473TC |
|||||||||
|
TT |
70 |
66 |
81 |
64.3 |
100 |
40.0 - 200.0 |
|||
|
TC + CC |
36 |
34 |
45 |
35.7 |
1.08 |
0.63 - 1.86 |
160 |
75.0 - 225.0 |
0.074 |
*Numbers may not sum 236 tumor specimens in cases of missing data. Abbreviations: HER-2: Human epidermal growth factor receptor 2.
Table 1: Distribution of clinical features and PTGS2 genotypes according to COX-2 immunostaining (IRS or CS).
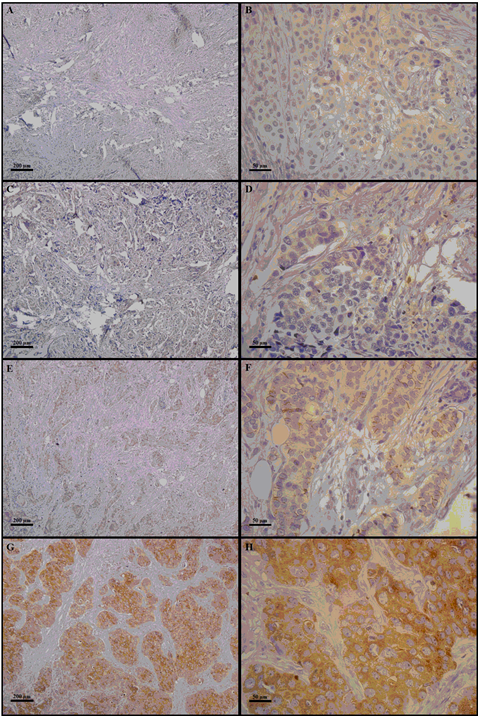
Figure 1: The right column indicates 100× magnification, and the left column indicates 400× magnification. Photomicrographs of COX-2 immunostaining in mammary tissue specimens from breast cancer patients. The panels (A–H) show different immunoreaction intensities: (A and B) negative; (C and D) weak; (E and F) moderate; (G and H) strong immunostaining.
Next, we evaluated if COX-2 protein levels in breast tumors, categorized as low or high according to IRS, could predict survival outcomes, but no significant effects were detected either on disease-free survival (HR=0.65; 95% CI 0.30-1.39) or overall survival (HR=1.10; 95% CI 0.39-2.83). In addition to evaluating the prognostic impact of COX-2 protein levels in the INCA-Brazil cohort, we decided to evaluate online available data on PTGS2 mRNA expression from other breast cancer cohorts compiled in the GEO, EGA and TCGA platforms [22]. Table 2 shows the impacts on relapse-free survival or overall survival, either considering all tumors together or stratifying cases into ER-positive, ER-negative, HER2-positive or basal-like. The results indicate significant protective effect of high COX-2 expression for both relapse-free and overall survival when considering all tumors or only ER-positive tumors. In contrast, within basal-like tumors, high COX-2 expression appears to contribute for worse survival outcomes, although the effect was only significant for relapse-free survival.
|
Tumor subset |
Probe |
Relapse-free survival |
Overall survival |
||||||
|
N |
HR |
95% CI |
P |
N |
HR |
95% CI |
P |
||
|
All tumors |
204748_at |
3951 |
0.82 |
0.72-0.92 |
0.001 |
1402 |
0.74 |
0.57-0.96 |
0.024 |
|
ER + |
204748_at |
2061 |
0.62 |
0.49-0.79 |
6.8 e-05 |
548 |
0.45 |
0.27-0.77 |
0.0026 |
|
ER - |
204748_at |
801 |
1.22 |
0.93-1.59 |
0.15 |
251 |
0.75 |
0.44-1.27 |
0.29 |
|
Basal-like |
204748_at |
618 |
1.6 |
1.22-2.11 |
0.0007 |
241 |
1.66 |
0.92-2.98 |
0.087 |
|
HER2 + |
204748_at |
252 |
0.79 |
0.51-1.23 |
0.3 |
129 |
0.5 |
0.23-1.08 |
0.074 |
Abbreviations: ER+: Estrogen receptor positive, ER-: Estrogen receptor negative, HER-2: Human epidermal growth factor receptor 2.
Table 2: Prognostic impact of high tumor levels of PTGS2 mRNA considering online data from compiled breast cancer cohorts [22].
4. Discussion
The present study aimed to investigate if PTGS2 SNPs could explain the variability on COX-2 expression in breast tumors, and to evaluate if the differential tumor expression of COX-2 would affect survival outcomes of breast cancer patients. The first approach was based on a single cohort of Brazilian breast cancer patients who had been previously genotyped for the PTGS2 SNPs [19] and had availability of tumor blocks. Individual information was used to select tumor specimens from patients with variant genotypes, especially rs689466, which has been shown to increase gene transcription in different cell models [17, 18, 20]. The other three most frequent PTGS2 SNPs composing the major five haplotypes [27] were also present in the selection and could be simultaneously evaluated. The second approach was based on a compilation of publically available information regarding gene expression profiles, based on tumor mRNA levels, and survival outcomes of other breast cancer cohorts that are included in the GEO, EGA and TCGA platforms [22].
The results of COX-2 immunostaining within the INCA-Brazil cohort confirm the expected large variability on COX-2 expression in breast tumors [10], and indicate no significant effect of PTGS2 SNPs either considering the distribution of continuous imunohistochemistry scores or the proportion of tumors with high IRS (≥ 6). Although the lack of detectable associations affecting COX-2 expression could be attributable to be a type 2 error, due to the relatively limited sample size, the results regarding PTGS2 SNPs suggest small effects, if any. In contrast, high COX-2 scores were significantly associated with high-grade (G3) tumors, which seems in accordance with the expected roles of COX-2 and PGE2 in favoring tumor proliferation [28]. However, no significant prognostic impact was found regarding high COX-2 scores in breast tumors of the INCA-Brazil cohort. Few previous studies investigated the impact of high COX-2 imunostaining scores in breast cancer survival outcomes, considering at least 100 tumor specimens and a five-year follow-up [29-33]. Among those studies, only Siking et al. [31] reported a significant prognostic association, i.e. that high COX-2 scores in breast tumors (N=193) increased the risk of distant metastasis after multivariate analysis (HR=2.8, 95% CI 1.6-4.9; P=0.002).
In order to expand the evaluation of the potential prognostic impact of COX-2 expression in breast tumors, we considered publically available data on tumor mRNA from large breast cancer cohorts. The analysis of a large number of cases increases statistical power and allows stratification according to breast cancer subsets that might be differently associated with and/or affected by COX-2 expression. Indeed, the results indicate a significantly protective effect of high COX-2 mRNA for both relapse-free and overall survival when considering all tumors or only ER-positive tumors, whereas patients with basal-like tumors appear to have worse relapse-free survival when tumor expression of COX-2 is high. Such difference between basal-like and ER-positive tumors regarding COX-2 prognostic effect might be related to the superior ability of basal-like tumors in recruiting macrophages [34, 35] and inducing M2 polarization. This scenario involves higher COX-2 synthesis, and thereby favors protumorigenic functions [34, 36-38], such as epithelial–mesenchymal transition, proliferation, chemoresistance and motility of cancer cells [39]. In contrast, luminal tumors present lower macrophage infiltration, which is inversely related to ER positivity [40]. Moreover, it has been shown that moderate levels of M1 macrophages contribute to lower risk of relapse ER-positive disease [41].
5. Conclusions
Taken together, the results indicate that gene polymorphisms do not account for the variability on COX-2 protein levels in breast tumors, and that COX-2 mRNA may be a better candidate for prognostic evaluation of breast cancer survival, whose impacts depend on breast cancer subtypes. The disparities regarding the prognostic impact of COX-2 mRNA in breast cancer subtypes are likely to be associated with gene signatures, and reinforce the need for large and combined evaluations, so that all factors influencing breast cancer prognosis can be better evaluated.
6. Highlights
- Breast cancer is very heterogeneous, and new prognostic biomarkers are needed.
- Chronic inflammation affects carcinogenesis, and COX-2 is a major trigger.
- Gene polymorphisms affect gene transcription, but not protein level of tumor COX-2.
- The prognostic impact of tumor COX-2 mRNA depends on breast cancer subtypes.
- High COX-2 mRNA indicates higher risk of relapse of basal-like breast cancer.
Funding
The study was supported by Conselho Nacional de Pesquisa e Desenvolvimento (CNPq 310580/2018-8), Fundação Carlos Chagas Filho de Amparo à Pesquisa no Rio de Janeiro (FAPERJ E-26/210.784/2015 and E-26/200.015/2019), and by Coordenação de Apoio ao Pessoal de Nível Superior (CAPES) via Programa de Pós-Graduação em Saúde Pública e Meio Ambiente (ENSP-FIOCRUZ). DRF-A and JBP received schollarships from CAPES and CNPq, respectively.
Acknowledgements
The authors thank Matheus Andrade Rajão for taking pictures of the tumor slides and the personnel from the Breast Cancer Hospital (HC3-INCA), from the National Bank of Tumors in the Brazilian National Cancer Institute (BNT-INCA), and from Division of Pathology (DIPAT-INCA) for logistic support in sample and data collection.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Author Contribution Statement
Freitas-Alves DR: recruited patients, collected clinical and histopathological data, performed statistical analyses, generated tables and figures, and drafted the manuscript. Pinto JB: helped with statistical analyses and with design of figures and tables. Rodrigues FR: coordinated pathological evaluation of surgical resections, selected patients’ blocks and slides, attributed immunohistochemistry scores. Valverde P and Fernandes DCS: conducted immunohistochemical analyses. Accioly MT, Valença SS and Perini JA: contributed to the study rationale, data analyses and interpretation. Vianna-Jorge: designed and coordinated the study, analyzed the data, wrote and revised the manuscript. All authors read and approved the final manuscript.
References
- Torre LA, Islami F, Siegel RL, et al. Global Cancer in Women: Burden and Trends. Cancer Epidemiol Biomarkers Prev 26 (2017): 444-457.
- Dai X, Xiang L, Li T, Bai Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. J Cancer 7 (2016): 1281-1294.
- Bettaieb A, Paul C, Plenchette S, et al. Precision medicine in breast cancer: reality or utopia? J Transl Med 15 (2017).
- Ferlay J, Shin H-R, Bray F, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer J Int Cancer 127 (2010): 2893-2917.
- Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin Cancer Biol 52 (2018): 56-73.
- Elinav E, Nowarski R, Thaiss CA, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 13 (2013): 759-771.
- Aaltomaa S, Lipponen P, Eskelinen M, et al. Lymphocyte infiltrates as a prognostic variable in female breast cancer. Eur J Cancer 28 (1992): 859-864.
- Bahiraee A, Ebrahimi R, Halabian R, et al. The role of inflammation and its related microRNAs in breast cancer: A narrative review. J Cell Physiol 234 (2019): 19480-19493.
- Harris RE. Cyclooxygenase-2 and the inflammogenesis of breast cancer. World J Clin Oncol 5 (2014): 677.
- Glover JA, Hughes CM, Cantwell MM, Murray LJ. A systematic review to establish the frequency of cyclooxygenase-2 expression in normal breast epithelium, ductal carcinoma in situ, microinvasive carcinoma of the breast and invasive breast cancer. Br J Cancer 105 (2011): 13-17.
- Tanabe T, Tohnai N. Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat 68 (2002): 95-114.
- Appleby SB, Ristimäki A, Neilson K, et al. Structure of the human cyclo-oxygenase-2 gene. Biochem J 302 (1994): 723-727.
- Moore AE, Young LE, Dixon DA. A common single-nucleotide polymorphism in cyclooxygenase-2 disrupts microRNA-mediated regulation. Oncogene 31 (2012): 1592-1598.
- Zhang X, Miao X, Tan W, et al. Identification of functional genetic variants in cyclooxygenase-2 and their association with risk of esophageal cancer. Gastroenterology 129 (2005): 565-576.
- Piranda DN, Festa-Vasconcellos JS, Amaral LM, et al. Polymorphisms in regulatory regions of cyclooxygenase-2 gene and breast cancer risk in Brazilians: a case-control study. BMC Cancer 10 (2010): 613.
- Agundez J, Gonzalez-Alvarez D, Vega-Rodriguez M, et al. Gene Variants and Haplotypes Modifying Transcription Factor Binding Sites in the Human Cyclooxygenase 1 and 2 (PTGS1 and PTGS2) Genes. Curr Drug Metab 15 (2014): 182-195.
- Pereira C, Sousa H, Ferreira P, et al. COX-2 polymorphism may be a susceptibility marker for gastric adenocarcinoma in patients with atrophy or intestinal metaplasia. World J Gastroenterol 12 (2006): 5473-5478.
- Sakaki M, Makino R, Hiroishi K, et al. Cyclooxygenase-2 gene promoter polymorphisms affect susceptibility to hepatitis C virus infection and disease progression: SNP of COX-2 gene protects against HCV infection. Hepatol Res 40 (2010): 1219-1226.
- Freitas-Alves DR, Vieira-Monteiro H de A, Piranda DN, et al. PTGS2 polymorphism rs689466 favors breast cancer recurrence in obese patients. Endocr Relat Cancer ERC-17-0374 (2018).
- Piranda DN, Abreu RBV, Freitas-Alves DR, et al. Modulation of the prostaglandin-endoperoxide synthase 2 gene expression by variant haplotypes: influence of the 3′-untranslated region. Braz J Med Biol Res 51 (2018): e6546.
- Crespi E, Bottai G, Santarpia L. Role of inflammation in obesity-related breast cancer. Curr Opin Pharmacol 31 (2016): 114-122.
- Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123 (2010): 725-731.
- Vieira-Monteiro H de A, Freitas-Alves DR, Sobral-Leite M, et al. Prognostic evaluation of VEGFA genotypes and haplotypes in a cohort of Brazilian women with non metastatic breast cancer. Cancer Biol Ther 17 (2016): 674-683.
- Delou JM de A, Vignal GM, Índio-do-Brasil V, et al. Loss of constitutive ABCB1 expression in breast cancer associated with worse prognosis. Breast Cancer Targets Ther 9 (2017): 415-428.
- Surowiak P, Materna V, Matkowski R, et al. Relationship between the expression of cyclooxygenase 2 and MDR1/P-glycoprotein in invasive breast cancers and their prognostic significance. Breast Cancer Res 7 (2005): R862.
- Kirkegaard T, Edwards J, Tovey S, et al. Observer variation in immunohistochemical analysis of protein expression, time for a change? Histopathology 48 (2006): 787-794.
- Festa-Vasconcellos JS, Piranda DN, Amaral LM, et al. Polymorphisms in cycloxygenase-2 gene and breast cancer prognosis: association between PTGS2 haplotypes and histopathological features. Breast Cancer Res Treat 132 (2012): 251-258.
- Cao Y, Prescott SM. Many actions of cyclooxygenase-2 in cellular dynamics and in cancer. J Cell Physiol 190 (2002): 279-286.
- Ristimäki A, Sivula A, Lundin J, et al. Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res 62 (2002): 632-635.
- Sivula A, Talvensaari-Mattila A, Lundin J, et al. Association of cyclooxygenase-2 and matrix metalloproteinase-2 expression in human breast cancer. Breast Cancer Res Treat 89 (2005): 215-220.
- Zerkowski MP, Camp RL, Burtness BA, et al. Quantitative Analysis of Breast Cancer Tissue Microarrays Shows High Cox-2 Expression Is Associated with Poor Outcome. Cancer Invest 25 (2007): 19-26.
- Sicking I, Rommens K, Battista MJ, et al. Prognostic influence of cyclooxygenase-2 protein and mRNA expression in node-negative breast cancer patients. BMC Cancer 14 (2014): 952.
- Giaginis C, Sampani A, Kotta-Loizou I, et al. Elevated Hu-Antigen Receptor (HuR) Expression is Associated with Tumor Aggressiveness and Poor Prognosis but not with COX-2 Expression in Invasive Breast Carcinoma Patients. Pathol Oncol Res 24 (2018): 631-640.
- Sousa S, Brion R, Lintunen M, et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res 17 (2015): 101.
- Espinoza JA, Jabeen S, Batra R, et al. Cytokine profiling of tumor interstitial fluid of the breast and its relationship with lymphocyte infiltration and clinicopathological characteristics. OncoImmunology 5 (2016): e1248015.
- Stewart DA, Yang Y, Makowski L, Troester MA. Basal-like Breast Cancer Cells Induce Phenotypic and Genomic Changes in Macrophages. Mol Cancer Res 10 (2012): 727-738.
- Su S, Liu Q, Chen J, et al. A Positive Feedback Loop between Mesenchymal-like Cancer Cells and Macrophages Is Essential to Breast Cancer Metastasis. Cancer Cell 25 (2014): 605-620.
- Hollmén M, Roudnicky F, Karaman S, Detmar M. Characterization of macrophage - cancer cell crosstalk in estrogen receptor positive and triple-negative breast cancer. Sci Rep 5 (2015): 9188.
- Prasmickaite L, Tenstad EM, Pettersen S, et al. Basal-like breast cancer engages tumor-supportive macrophages via secreted factors induced by extracellular S100A4. Mol Oncol 12 (2018): 1540-1558.
- Garvin S, Vikhe Patil E, Arnesson L-G, et al. Differences in intra-tumoral macrophage infiltration and radiotherapy response among intrinsic subtypes in pT1-T2 breast cancers treated with breast-conserving surgery. Virchows Arch 475 (2019): 151-162.
- Ali HR, Chlon L, Pharoah PDP, et al. Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLOS Med 13 (2016): e1002194.


 Impact Factor: * 4.1
Impact Factor: * 4.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 