Effect of Point Mutations in SOD1 Enzyme Stability and Solubility: A Study Using Computational Approach
Article Information
Rutuja Yelmar#, Haifa Parkar#, Norine D’Souza*
St. Xavier’s College (Autonomous), Mumbai, India
*Corresponding Author: Norine D’Souza, St. Xavier’s College (Autonomous), Mumbai, India
# - Contributed Equally
Received: 15 July 2020; Accepted: 19 August 2020; Published: 21 August 2020
Citation: Rutuja Yelmar, Haifa Parkar, Norine D’Souza. Effect of Point Mutations in SOD1 Enzyme Stability and Solubility: A Study Using Computational Approach. Journal of Bioinformatics and Systems Biology 3 (2020): 058-073.
View / Download Pdf Share at FacebookAbstract
Structural integrity and functional properties of protein are governed by arrangement and side-chain nature of every amino acid constituting it. Substitution mutations have been observed in many proteins which are very important for cellular functions. The current study aims to understand the molecular basis of protein stability, and solubility using SOD1 enzyme where multiple point mutations are have been reported. Around 51 point mutations reported in the SOD1 enzyme were retrieved from ALSoD and HuVarBase databases. The mutations were grouped based on the properties of amino acid residue involved. The impact of mutations on SOD1 function was predicted using PROVEAN and changes in the structural stability induced by point mutation was evaluated based on free energy change using Foldx. Influence of mutation on SOD1 solubility was studied using CamSol Intrinsic web server. The mutations resulting from the substitution of polar and charged residues were found to influence functionally important state, free energy value and solvent interaction of the protein. This study hypothesizes that the physicochemical properties of every amino acid residue constituting protein, influence the stability and solubility of the protein. The future prospect includes establishing a correlation between these two properties to identify factors driving protein aggregation upon point mutation.
Keywords
Stability; Solubility; Free energy; SOD1; Physicochemical properties
Stability articles, Solubility articles, Free energy articles, SOD1 articles, Physicochemical properties articles
Stability articles Stability Research articles Stability review articles Stability PubMed articles Stability PubMed Central articles Stability 2023 articles Stability 2024 articles Stability Scopus articles Stability impact factor journals Stability Scopus journals Stability PubMed journals Stability medical journals Stability free journals Stability best journals Stability top journals Stability free medical journals Stability famous journals Stability Google Scholar indexed journals Solubility articles Solubility Research articles Solubility review articles Solubility PubMed articles Solubility PubMed Central articles Solubility 2023 articles Solubility 2024 articles Solubility Scopus articles Solubility impact factor journals Solubility Scopus journals Solubility PubMed journals Solubility medical journals Solubility free journals Solubility best journals Solubility top journals Solubility free medical journals Solubility famous journals Solubility Google Scholar indexed journals Free energy articles Free energy Research articles Free energy review articles Free energy PubMed articles Free energy PubMed Central articles Free energy 2023 articles Free energy 2024 articles Free energy Scopus articles Free energy impact factor journals Free energy Scopus journals Free energy PubMed journals Free energy medical journals Free energy free journals Free energy best journals Free energy top journals Free energy free medical journals Free energy famous journals Free energy Google Scholar indexed journals SOD1 articles SOD1 Research articles SOD1 review articles SOD1 PubMed articles SOD1 PubMed Central articles SOD1 2023 articles SOD1 2024 articles SOD1 Scopus articles SOD1 impact factor journals SOD1 Scopus journals SOD1 PubMed journals SOD1 medical journals SOD1 free journals SOD1 best journals SOD1 top journals SOD1 free medical journals SOD1 famous journals SOD1 Google Scholar indexed journals Physicochemical properties articles Physicochemical properties Research articles Physicochemical properties review articles Physicochemical properties PubMed articles Physicochemical properties PubMed Central articles Physicochemical properties 2023 articles Physicochemical properties 2024 articles Physicochemical properties Scopus articles Physicochemical properties impact factor journals Physicochemical properties Scopus journals Physicochemical properties PubMed journals Physicochemical properties medical journals Physicochemical properties free journals Physicochemical properties best journals Physicochemical properties top journals Physicochemical properties free medical journals Physicochemical properties famous journals Physicochemical properties Google Scholar indexed journals amino acid articles amino acid Research articles amino acid review articles amino acid PubMed articles amino acid PubMed Central articles amino acid 2023 articles amino acid 2024 articles amino acid Scopus articles amino acid impact factor journals amino acid Scopus journals amino acid PubMed journals amino acid medical journals amino acid free journals amino acid best journals amino acid top journals amino acid free medical journals amino acid famous journals amino acid Google Scholar indexed journals HuVarBase databases articles HuVarBase databases Research articles HuVarBase databases review articles HuVarBase databases PubMed articles HuVarBase databases PubMed Central articles HuVarBase databases 2023 articles HuVarBase databases 2024 articles HuVarBase databases Scopus articles HuVarBase databases impact factor journals HuVarBase databases Scopus journals HuVarBase databases PubMed journals HuVarBase databases medical journals HuVarBase databases free journals HuVarBase databases best journals HuVarBase databases top journals HuVarBase databases free medical journals HuVarBase databases famous journals HuVarBase databases Google Scholar indexed journals PROVEAN articles PROVEAN Research articles PROVEAN review articles PROVEAN PubMed articles PROVEAN PubMed Central articles PROVEAN 2023 articles PROVEAN 2024 articles PROVEAN Scopus articles PROVEAN impact factor journals PROVEAN Scopus journals PROVEAN PubMed journals PROVEAN medical journals PROVEAN free journals PROVEAN best journals PROVEAN top journals PROVEAN free medical journals PROVEAN famous journals PROVEAN Google Scholar indexed journals SOD1 articles SOD1 Research articles SOD1 review articles SOD1 PubMed articles SOD1 PubMed Central articles SOD1 2023 articles SOD1 2024 articles SOD1 Scopus articles SOD1 impact factor journals SOD1 Scopus journals SOD1 PubMed journals SOD1 medical journals SOD1 free journals SOD1 best journals SOD1 top journals SOD1 free medical journals SOD1 famous journals SOD1 Google Scholar indexed journals
Article Details
1. Introduction
The structural dynamics of protein has been mainly governed mainly by its stability and solubility. However, these two properties are in turn influenced by amino acid residues with distinct physicochemical properties attributed to side-chain groups which are vital for maintaining the structural and functional integrity [1, 2]. At physiological pH 7.0, the side-chain group of amino acids are charged or uncharged depending on their pK values, which determines their solvent accessibility. The residues with charged groups like lysine (K), glutamine (Q) have more solvent accessibility as they are less buried; whereas residues like tyrosine(Y), isoleucine (I) with uncharged side-chains have less solvent accessibility and are more buried [3].
The ionized side-chain of amino acid residue in a protein contributes to attractive or repulsive interaction between neighboring charged surface residues and buried ionizable, carboxyl groups contribute to the stability of the protein by hydrogen and other interactions [4, 5]. Experimental work suggests the role of hydrogen bonding between amino acids carrying –OH group and side chains of neighboring amino acids as a contributing factor of protein stability [6]. Amino acids with ionizable side-chain groups, such as Aspartic acid (D), Glutamic acid (E) have shown to improve the solubility of the protein considerably [7]. The precise configuration of the protein as coded by the gene and structural organization of amino acids is essential for the functioning. The mutation of specific residues in certain proteins has been shown to promote protein aggregation, disrupt protein-protein interaction, and develop drug resistance. Such type of mutations has been considered as one of the contributing factors in the manifestation of neurodegenerative disorders such as Amyotrophic lateral sclerosis, Alzheimer’s disease, Parkinson’s disease, etc.[8]. Literature suggests the presence of multiple point mutations in SOD1 as one of the factors promoting the progression and pathogenesis of familial ALS [6]. ALSoD [9] and HuVarBase [10] databases provide point mutation data associated with SOD1. Although a lot of point mutations have been listed for SOD1 their exact impact on protein dynamics in terms of stability and solubility has been less explored [11]. In the present study, the impact of point mutations on protein function, stability and solubility has been analyzed using the SOD1 enzyme and thereby to study its potential association with protein aggregation leading to ALS as one of the possible factors.
2. Methodology
2.1 Predicting impact of mutations on SOD1 function
The point mutations for SOD1 were retrieved from ALSoD and HuVarBase and were classified based on the nature of mutated amino acid. The wild type (WT) SOD1 sequence was retrieved from UniprotKB (P00441). The impact of mutations on the function of SOD1 was predicted for WT and 51 mutants using PROVEAN (Protein Variation Effect Analyzer) tool [12].
2.2 Analyzing the effect of mutations on structural stability:
The structure of SOD1 (1PU0) was retrieved from protein data bank (https://www.rcsb.org/). All deleterious and neutral mutations predicted based on the Provean scores were subjected for free energy change (ΔΔG) calculations using the FoldX suite [13]. Prior to free energy calculation the WT structure was refurbished using the repair command. Interatomic clashes produced by the mutated residue with the residues in the vicinity were studied for selected mutants using the findClash function of UCSF chimera. Substitution mutations were induced in SOD1 using Dunbrack rotamer library [14].
2.3 Analyzing the effect of the mutation on solubility:
The impact of 51 point mutations on Solubility was predicted using CamSol Intrinsic web server. A solubility score for mutants above -1, when compared with WT, is considered to be more soluble while the score less than -1 is less soluble [15].
3. Result
3.1 Predicting impact of mutations on SOD1 function
All the point mutations reported in SOD1 protein were listed from disease-specific database ALSoD, HuVarBase. Based on the nature of the side-chain group of amino acids, mutations were classified as polar (charged and uncharged) and nonpolar. Categorization has been listed in Figure 1.a. This variation in the properties of the amino acid side-chain was explored to group mutated SOD1 variants. Most of the mutations resulted from the substitution of WT nonpolar residue with nonpolar ones. Further categorization of point mutant based on their origin has been supplied as supplement data (Supplementary Figure 1.b)
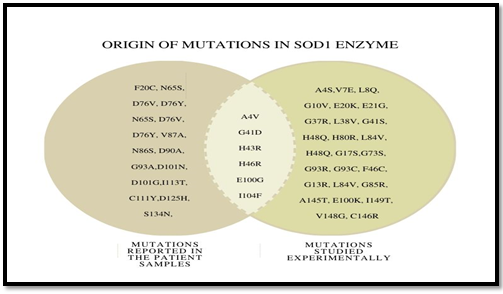
Figure 1.b: Origin of Mutations in SOD1
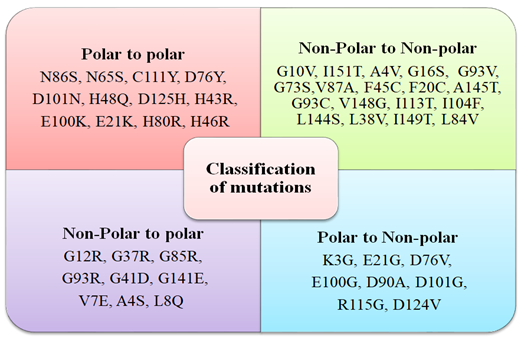
Figure 1.a: List of SOD1 mutations grouped into polar-polar, nonpolar-nonpolar, nonpolar-polar and polar-nonpolar (clockwise) categories based on the nature of amino acid residue side-chain.
The impact of point mutations on the function of SOD1 was predicted using Provean. Any mutation in sequence with provean score below -2.5 is considered to be deleterious for protein. The provean score obtained for each of the point mutations has been represented in Figure 2 a-d, For polar to polar point mutations it was observed that the provean score of mutations ranged between -1 to -8. H80R has the lowest value of -8 followed by D76Y wherein C111Y, E100K and E21K were shown to have value more than -2.5 indicating the neutral nature (Figure 2.a). For nonpolar to nonpolar point mutations, all the mutations were found to be deleterious in nature with G93V, G93C and F45C having the lowest provean score of -8.27, -8.02 and -7.51 respectively (Figure 2.b). Similar trend was observed for nonpolar to polar mutations with G85R having the lowest score of -7.91 followed by G37R, G141E and G93R (Figure 2.c). Polar to nonpolar SOD1 mutations found to be deleterious except for D90A (Figure 2.d).
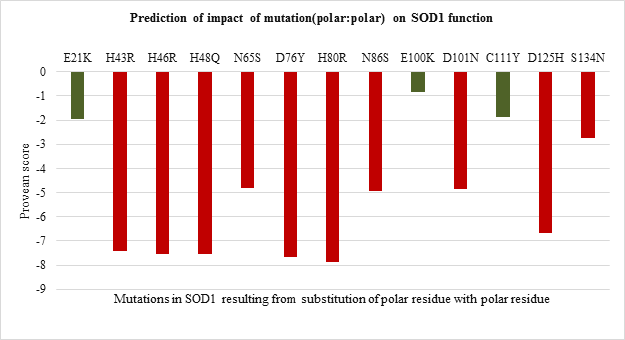
Figure 2.a: Effect of Polar: Polar substitution mutation on SOD1 enzyme function.
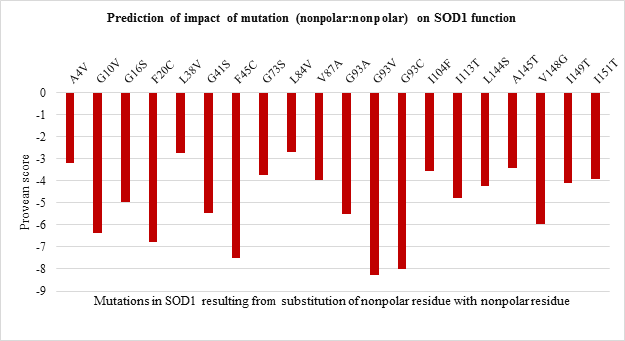
Figure 2.b: Effect of Nonpolar: Nonpolar substitution mutation on SOD1 enzyme function.
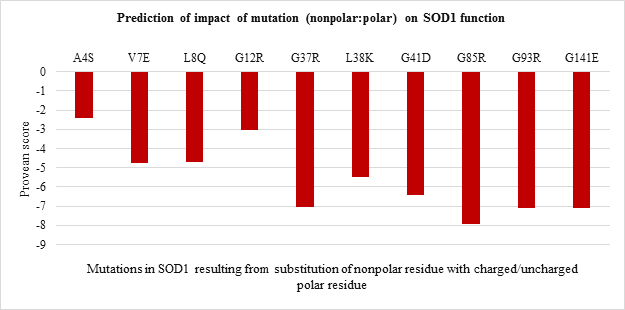
Figure 2.c: Effect of Nonpolar: Polar substitution mutation on SOD1 enzyme function.
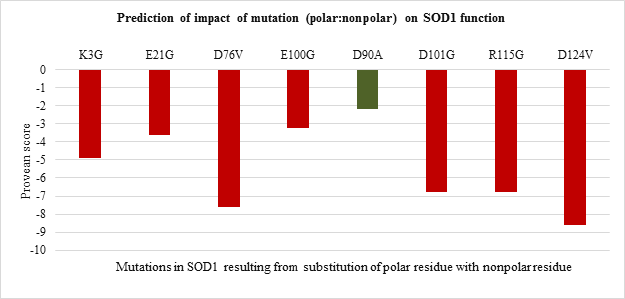
Figure 2.d: Effect of Polar: Nonpolar substitution mutation on SOD1 enzyme function.
3.2 Effect of point mutations on protein stability
Impact of mutation on protein stability was evaluated based on free energy change calculations and inter-atomic interactions. Among polar to polar SOD1 mutations (Figure 3.a) it was observed that H46R has highest ??G values followed by S134N and H80R and point mutations E21K, N65S, D76Y, E100K and D125H were found to be stabilizing in nature.
All the 20 nonpolar to nonpolar SOD1 mutations were found to be destabilizing wherein I104F, G10V, G16S were highly destabilizing while L144S mutation was found to be least destabilizing with the lowest ??G values. (Figure 3.b). Among nonpolar to polar and polar to nonpolar point mutations (Figure 3.c and Figure 3.d respectively) showed destabilizing impact. G37R, G85R and G141E in nonpolar to polar category were highly destabilizing (Figure 3.c) while D124V, D90A and D101G were highly destabilizing in polar to nonpolar category (Figure 3.d).
Furthermore, interatomic interactions between the side-chain of mutated residue and neighboring residues were analyzed and represented in the form of pseudobonds [16]. The yellow colored bonds correspond to steric clash caused by atoms of mutated residue unveiling basis for destabilization. All destabilizing mutations show steric clashes at varying degrees (Figures 4 a-h).
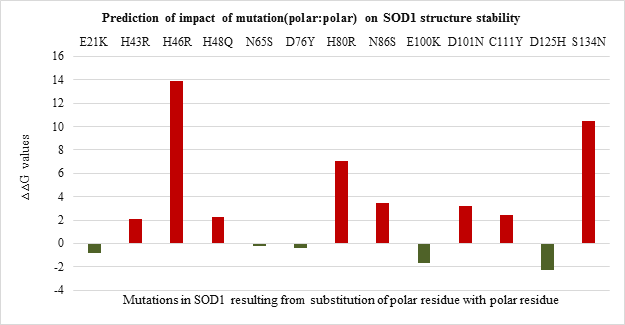
Figure 3.a: Effect of Polar: Polar substitution mutation on SOD1 enzyme structural stability.
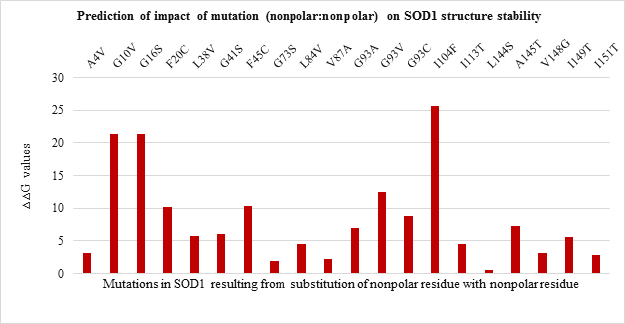
Figure 3.b: Effect of Nonpolar: Nonpolar substitution mutation on SOD1 enzyme structural stability.
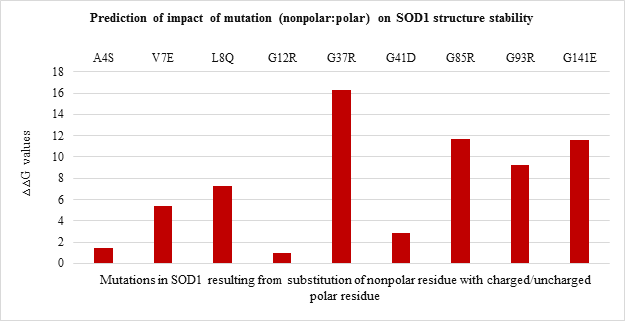
Figure 3.c: Effect of Nonpolar: Polar substitution mutation on SOD1 enzyme structural stability
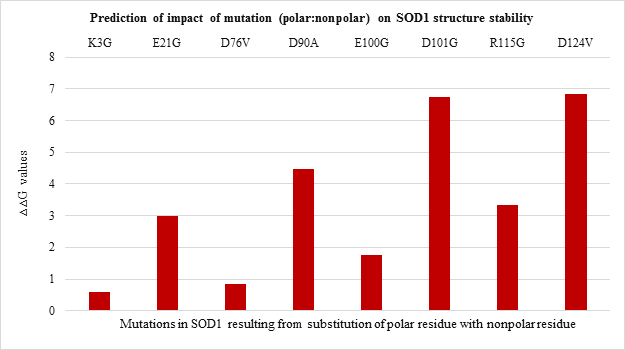
Figure 3.d: Effect of Polar: Nonpolar substitution mutation on SOD1 enzyme structural stability
Mutations resulting from the substitution of the same residues drastically varied in their free energy change values and thereby differing in destabilizing effect. The analysis of interatomic clashes between mutated residue and neighboring residue revealed the basis of destabilization. The relative position, property of side-chain of mutated residue, and nature of neighboring amino acid found to influence the number of clashes caused by the mutated residue which in turn affected the stability of SOD1. For example, the clashes created by mutant G12R are less than clashes created by mutant G37R as the angle of rotamer, its position and surrounding amino acid residue varies. (Figure 4. f and Figure 4.g)
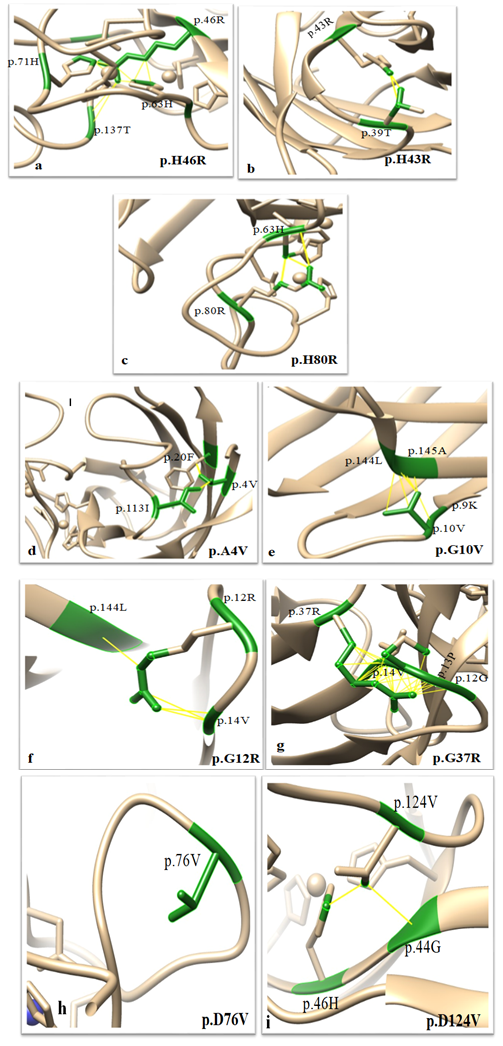
Figure 4: Clashes created by Side-chain of mutated residue with residues in the vicinity: a. The side-chain of p.46R making clashes with side-chains of p.63H, p137T and p.71H. b. The side-chain of p.43R making clashes with side chains of p.39T. c. The side-chain of p.80R making clashes with side-chains of p.63H. d. The side-chain of p.4V is making clashes with side-chains of p.20F and p.113I. e. The side-chain of p.10V is making clashed with the side-chains of p.144L and p.145A. f. The side-chain of p.12R is making clashes with side-chains of p.14V and p.144L.g. The side-chain of p.37R is making clashes with side-chains of p.12G, p.13P and p.14V. h. The side-chain of p.76V is not making any clash with the neighboring residues. i. The side-chain of p.124V making clashes with side-chains of p.44G and p.86N.
3.3 Analyzing effect of mutation on solubility
The solubility score prediction using CamSol Intrinsic showed a value of 1.70 for WT SOD1 which was used as a reference. The mutant SOD1 with solubility score below 1.70 are relatively less soluble while one with solubility score above 1.70 are relatively more soluble than WT SOD1. The solubility score predicted for 51 mutations is shown in Figure 5. a-d.
Amongst polar to polar residue substitution mutations, H46R and D76Y mutants were found to be more and less soluble compared to WT SOD1 respectively (Figure 5.a). While solubility of nonpolar to nonpolar residue substitution mutation was nearly the same as WT enzyme (Figure 5.b). Apart from this, all mutants resulting from nonpolar to polar residue substitution seemed to be more soluble than WT protein except G85R mutant (Figure 5.c). In contrast, all polar to nonpolar residue substitution mutants were relatively less soluble than WT SOD1 enzyme except E100G mutant (Figure 5.d).
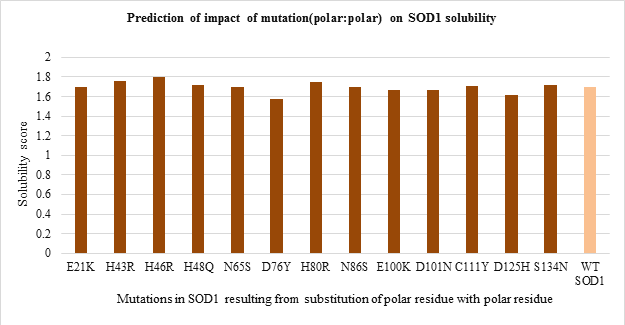
Figure 5.a: Effect of Polar: Polar substitution mutation on SOD1 enzyme solubility.
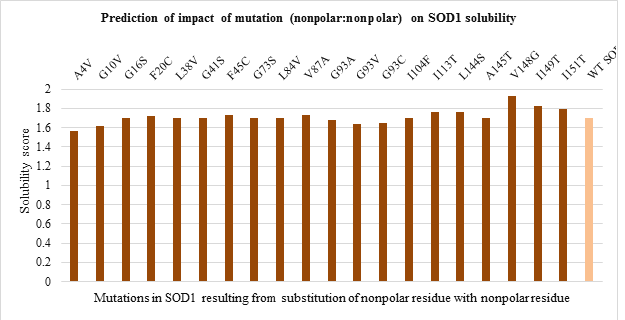
Figure 5.b: Effect of Nonpolar: Nonpolar substitution mutation on SOD1 enzyme solubility.
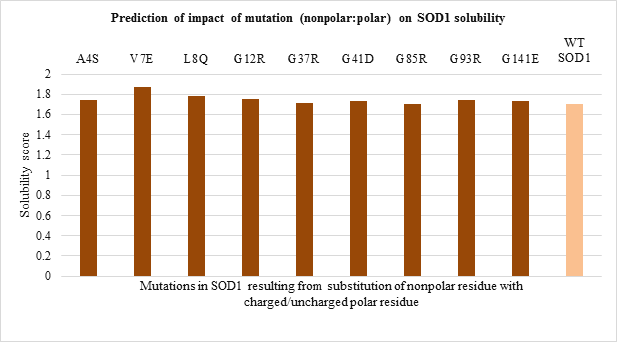
Figure 5.c: Effect of Nonpolar: Polar substitution mutation on SOD1 enzyme solubility.
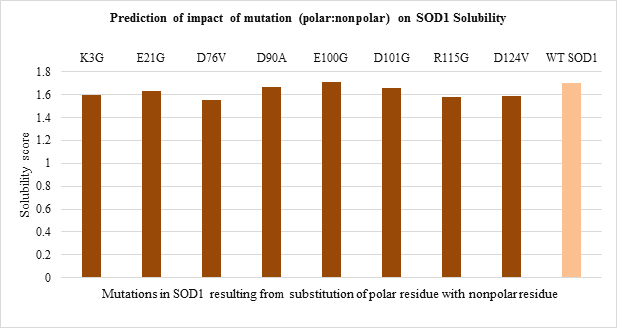
Figure 5.d: Effect of Polar: Nonpolar substitution mutation on SOD1 enzyme solubility.
4. Discussion
The structural and functional dynamics of protein is significantly influenced by the functional group of amino acids. Point mutations in proteins alter this dynamism which in turn depends upon the type of point mutation [17-20].
Thus the current study is an attempt to unveil the contribution of amino acid residues based on the nature of side-chain group, position, and interaction with neighboring residue in maintaining the structural stability and functional properties. The 51 SOD1 substitution mutations were selected from the databases. Majority of the mutations belong to nonpolar to nonpolar category followed by polar to polar category. Least number of mutations belong to polar to nonpolar category. Studies on impact prediction by provean analysis showed most of the mutants being deleterious. Irrespective of the nature of amino acid substituted, based on the provean score H80R, H48Q, H46R, H43R, D124V, D125H, G85R, G93V, G93C, G76Y are predicted to be more deleterious for SOD1 function. Among top 10 most deleterious mutations, H80R, H46R, H48Q and H43R are known to directly coordinate with Zn and Cu ions crucial for SOD1 activity, thus their substitution can impact SOD1 activity [21]. Whereas mutations G76Y, G85R, G93V, G93C, G124V, G125H resulted from the substitution of evolutionarily conserved residues can impact functional properties. Similar study was demonstrated by Zhang et al. 1992 where they reported no detectable or less activity of the human 5-lipoxygenase enzyme in which C-terminal conserved residues were mutated [22]. On the other hand, the neutral effect of E21K, D90A, E100K, C111Y mutants on SOD1 activity could be a result of the non-conserved nature of the residues that were mutated in the variants. The polarity, non-polarity, side-chain group bulkiness, and charge of amino acid residues seem to play a more important role over their conserved nature in case of structural stability of the SOD1 enzyme. The gain in the stability and relatively less destabilization of SOD1 resulting from the substitution of WT polar residue with another polar residue indicates they are vital for stability. In case of E100K, substitution of negatively charged glutamic acid (E) at 100th position with Lysine (K) has a stabilizing effect on the SOD1 structure. While substitution of positively charged histidine (H) with arginine (R) with the same charge has a less destabilizing effect than mutants from the same and other three (nonpolar to nonpolar, nonpolar to polar, polar to nonpolar) groups. Most of the mutations resulting from the substitution of the same polar residue with nonpolar residue and vice versa varied in their ??G values. Mutations like H80R, H46R, G37R, D124V were found to be more destabilizing which is apparently due to the difference in polarity and bulkiness of substituted residues whereas exceptions including H43R, G12R, D76V were found to be less destabilizing mutations. Inter-atomic clashes studied for the selected high and low destabilizing mutations resulting from substitution of same residues showed interaction with the neighboring residues. The analysis of inter-atomic interaction between mutated residue and side-chain residue in the nearest environment helps to identify the possible reason for SOD1 destabilization due to steric clashes. This study hypothesize, in addition to the physicochemical property of mutated residue, degree of freedom of rotamer selected, its relative position in three-dimensional space and nature of amino acid residues in vicinity governs the type of interactions mutated residue participate in. It was found that less destabilizing mutations create less steric clashes compared to high destabilizing ones. Considering the importance of the polarity of amino acid in structural stabilization of SOD1, its role in improving the solubility of the protein has been well studied for other proteins like RNase-Sa [17]. The current study also sheds light on the role of side-chain properties of amino acid residue and influence on protein solubility which can further influence its aggregation propensity [17, 20]. The solubility score determined by CamSol Intrinsic web server suggests that negatively and positively charged amino acids like aspartic acid, glutamic acid, glutamine, asparagine, arginine, and lysine can make mutant SOD1 more soluble than WT form [22-25]. Since mutants resulting from their substitution have less solubility score and mutant carrying them as substituent has a better solubility score. This observation holds true for all mutation groups. The key finding of the work suggests that polar / charged residues play an important role in maintaining the structural and functional dynamics of the protein. Furthermore, the present work opens up avenues to understand the effect of the accumulated mutation on the functional properties of the protein. Since the presence of SOD1 protein aggregates has been reported to play a role in the pathogenesis of familial ALS.
5. Conflict of Interest
The authors have no conflict of interest.
References
- Idicula-Thomas S and Balaj P V. Correlation between the Structural Stability and Aggregation Propensity of Proteins. In Silico Biol 7 (2007): 225-237.
- Pace C N, Grimsley G R, Scholtz J M. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem 284 (2009): 13285-9.
- Mason P E, Neilson G W, Dempsey C E, Barnes A C and Cruickshank J M. The hydration structure of guanidinium and thiocyanate ions: Implications for protein stability in aqueous solution. Proc. Natl. Acad. Sci 100 (2003): 4557–4561.
- Radivojac P, Iakoucheva L M, Oldfield C J, Obradovic Z, Uversky VN and Dunker A K. Intrinsic disorder and functional proteomics. Biophys J 92 (2007): 1439–1456.
- Pace C N, Fu H, Lee Fryar K, Landua J et al. Contribution of hydrogen bonds to protein stability. Protein Sci 23 (2014): 652–661
- Ross C. and Poirier M. Protein aggregation and neurodegenerative disease. Nat Med 10 (2004): S10–S17.
- Kramer R M, Shende V R, Motl N, Pace C N. & Scholtz J M. Toward a molecular understanding of protein solubility: increased negative surface charge correlates with increased solubility. Biophys journal 102 (2012): 1907–1915.
- Taylor J P, Brown R H Jr and Cleveland D W. Decoding ALS: from genes to mechanism. Nature 539 (2016): 197-206.
- ALSoD: Amyotrophic Lateral Sclerosis Online genetics Database. Available at: http://alsod.iop.kcl.ac.uk/home.aspx
- Ganesan K, Kulandaisamy A, Binny Priya S, Gromiha M M. HuVarBase: A human variant database with comprehensive information at gene and protein levels. PLoS ONE 14 (2019): e0210475.
- Ciryam P, Lambert-Smith I A, Bean D M, Freer R, Cid F, Tartaglia G G, Saunders D N, Wilson M R, Oliver S G, Morimoto R I. and Dobson CM. Spinal motor neuron protein supersaturation patterns are associated with inclusion body formation in ALS. Proc Natl Acad Sci 114 (2017): E3935-E3943.
- Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31 (2015): 2745-2747.
- Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res 33 (2005) (Web Server issue): W382–W388.
- Shapovalov M S, and Dunbrack R L. Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure 19 (2011): 844-858
- Sormanni P, Aprile F A. and Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol 427 (2015): 478-490.
- Vijaykumar M, Wong KY, Schreiber G, Fersht AR, Szabo A and Zhou HX. Electrostatic enhancement of diffusion-controlled protein-protein association: comparison of theory and experiment on barnase and barstar. J Mol Biol 278 (1998): 1015–1024.
- Trevino S R, Scholtz J M. & Pace C N. Amino acid contribution to protein solubility: Asp, Glu, and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J. Mol. Biol. 366 (2007): 449–460.
- Zayas J F. Solubility of Proteins. In: Functionality of Proteins in Food. Springer (1997).
- McGowen J C, and Mellors A. Relationships between the solubility of amino acids. J Appl Biochem 1 (1979): 423.
- Hayakawa S. and Nakai S. Relationships of hydrophobicity and net charge to the solubility of milk and soy proteins. J Food Sci 50 (1985): 486
- Wang J, Xu G, Gonzales V, et al. Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol Dis 10 (2002): 128-138.
- Zhang Y Y, Radmark O, and Samuelsson B. Biochemistry Mutagenesis of some conserved residues in human 5-lipoxygenase: Effects on enzyme activity. Proc Nati Acad Sci 89 (1992): 485-489.
- Kramer R M, Shende V R, Motl N, Pace C N. & Scholtz J M. Toward a molecular understanding of protein solubility: increased negative surface charge correlates with increased solubility. Biophys journal 102 (2012): 1907–1915
- Smialowski P, Doose G, Torkler P, Kaufmann S. & Frishman D. Proso II–a new method for protein solubility prediction. FEBS J 279 (2012): 2192-2200
- Niwa T, Ying B W, Saito K, et al. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc Natl Acad Sci 106 (2009): 4201–4206.


 Impact Factor: * 4.2
Impact Factor: * 4.2 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 