Enterotoxigenic Escherichia Coli Toxins and Colonization Factors Among Zambian Children Presenting with Moderate to Severe Diarrhea to Selected Health Facilities
Article Information
Michelo Simuyandi*1, Roma Chilengi1, Sean B Connor2, Joseph B. Voeglein2, Natasha M. Laban1, Katayi Mwila-Kazimbaya1, Caroline C. Chisenga1, John Mwaba1, David A. Sack2, Subhra Chakraborty2
1Enteric Disease and Vaccine Research Unit, Centre for infectious Disease Research in Zambia, Zambia
2Department of International Health, Johns Hopkins University, Baltomore, USA
*Corresponding author: Michelo Simuyandi, Enteric Disease and Vaccine Research Unit, Centre for Infectious Disease Research in Zambia, Lusaka, Zambia
Received: 08 July 2019; Accepted: 04 November 2019; Published: 11 November 2019
Citation:
Michelo Simuyandi, Roma Chilengi1, Sean B Connor, Joseph B. Voeglein, Natasha M. Laban, Katayi Mwila-Kazimbaya, Caroline C. Chisenga, John Mwaba, David A. Sack, Subhra Chakraborty. Enterotoxigenic Escherichia coli Toxins and Colonization factors among Zambian Children Presenting with Moderate to Severe Diarrhea to Selected Health Facilities. Archives of Microbiology & Immunology 3 (2019): 173-184.
View / Download Pdf Share at FacebookAbstract
Enterotoxigenic Escherichia Coli (ETEC) is an important cause for diarrheal disease in children and travelers globally. Epidemiological data on the distribution of strains of ETEC and associated colonization factors (CFs) is important for evaluation of candidate vaccines. We used conventional PCR and quantitative PCR to screen for toxins and CFs using DNA extracted from stool samples which tested positive for ETEC using the Luminex GPP panel collected from children presenting with moderate to severe diarrhea at selected health facilities in Lusaka. 49/106 (46.2%) were positive for at least one toxin (i.e. LT/STh/STp), ST was 18 (17%) [STh 16(15%) and STp 2 (~2%)], and LT 16(15%). The most frequent CF detected was CS6 with 6/49 (12.2%), followed by CS2, CS3 and CS7 with 2/49 (4.1%) each. CS6 was common across all toxin combinations (LT only, STh only and a combination of LT/STh) while CS2, CS3, CS7 were identified in both LT and LT/STh strains respectively. The mean age of children with detected toxin or CFs was 15.4 months (95% CI: 12.2, 18.7). Our results offer an insight into relevant CFs in ETEC diarrhea in Zambia and that Luminex™ platform is not as specific as ordinary and quantitative PCR for ETEC detection.
Keywords
Enterotoxigenic Escherichia coli; Zambia, Diarrhea; Colonization factors (CFs); Toxins; Children
Enterotoxigenic Escherichia Coli articles, Zambia articles, Diarrhea articles, Colonization factors (CFs) articles, Toxins articles, Children articles
Article Details
Introduction
Enterotoxigenic Escherichia coli (ETEC) is one of the major causes of bacterial diarrhea in low and middle-income countries (LMICs), especially among children and travelers to these regions. ETEC is estimated to cause about 1.7 billion annual episodes of diarrheal illness in LMIC countries lacking access to clean water and basic sanitation [1–3].
Even though the overall death rate from acute diarrheal illness has declined in the past few decades [4], morbidity remains unacceptably high. Repeated bouts of diarrhea disease have been linked to cognitive and developmental impairment, life-long disability, increased risk of cardiovascular disease later in life and reduced life expectancy, and ETEC particularly contributes substantially to post diarrheal sequelae that follow repeated enteric infections [5, 6]. There are approximately more than 100 million children who are thought to suffer from gut dysfunction and its associated morbidities of stunted growth, malnutrition, poor response to oral vaccines, and impaired cognitive development currently [6, 7]. This has led to an increase in the drive to develop vaccines for key diarrheal etiological pathogens including ETEC [8].
ETEC produces enterotoxins and colonization factors (CFs) - proteinaceous surface polymers that facilitate adherence to the intestinal mucosa. More than 25 different CFs have been identified, classified into different families including: (I) CFA/I-like group which include CFA/I, CS1, CS2, CS4, CS14, and CS17; (ii) a CS5-like group which comprise CS5, CS7, CS18 and CS20; and (iii) Class1b which has CS3, CS6, CS10, CS11 and CS12 [9]. The most prevalent CFs are CFA/I, CS1-CS6 and CS21; however more than 30% of all clinical ETEC isolates lack a known CF [10]. To cause diarrhea, ETEC bacteria must adhere to small bowel enterocytes [8], a process mediated by one or more CFs [11, 12]. Once colonized, ETEC strains elaborate heat-labile toxins (LT) and/or heat-stable toxins (ST) that disrupt fluid homeostasis, leading to fluid hyper-secretion and watery diarrhea. The general approach to ETEC vaccines involves the inclusion of most common colonization factors CFs with an heat-labile enterotoxin (LT) component to provide broad coverage [8, 13]. Therefore, defining the epidemiology of CFs in high burden populations has important implications for current ETEC vaccine candidates. Current advanced candidates include inactivated strains overexpressing CFA/I, CS3, CS5, and CS6 mixed with an LT B-subunit related toxoid and adjuvant (ETVAX vaccine) [14], and a live attenuated ETEC vaccine that expresses CFA/I, CS1, CS2, CS3, CS5, CS6 with the B subunit of LT (ACE527 vaccine) [15].
One candidate vaccine, ETVAX vaccine, a killed oral vaccine consisting of E coli expressing different CFAs, is reported to have induced strong mucosal (faucal and antibody in lymphocyte supernatant, ALS) IgA responses to all vaccine associated CFs and LTB, and memory responses to these antigens remained for at least 13–23 months [8, 16, 17].
In this report, we describe the presence and distribution of toxins and the circulating CFs in children presenting with moderate and severe diarrhea in Lusaka province, Zambia as a potential future evaluation site for ETEC, Shigella and other enteric vaccines.
Methods
Study design
The stool samples used in this study were obtained from a study that evaluated rotavirus vaccine effectiveness in Zambian infants presenting with moderate to severe diarrhea (MSD) at health facilities. The design of the study is described in detail elsewhere [18, 19]. In this paper we report the distribution of toxins and CFs among the ETEC positive samples based on the ordinary PCR and the quantitative PCR. Diarrhea was defined by caregiver confirmation of child having ≥3 abnormally loose stools in the past 24 hrs.
Nucleic acid extraction
The Nucleic acid extraction was performed according to [18] were stored at -20 °C before shipment.
Detections of enterotoxins and colonization factors
DNA samples positive for ETEC by Luminex™ xTag were screened for ETEC toxins via conventional PCR using the method described by Rodas et al. 2011 [20] after modifications. The final concentrations of the reaction mixes were 1X DreamTaq Green buffer, 0.625 U DreamTaq polymerase, 1.2 mM dNTPs (300 μM each, Denville Scientific, MA), 0.2 μM LT-F primer, 0.2 μM LT-R primer, 0.4 μM STp-F primer, 0.4 μM STp-R primer, 0.9 μM STh-F primer, 0.9 μM STh-R primer, and 4.0 μL (approximately 100 ng) of sample DNA, brought to 25 μL final volume using Ultrapure distilled water (Invitrogen, CA). The primer sequences are given in Table 1. The reactions were performed in 8-well strips (Bio-Rad, CA) using SimpliAmp Thermocycler (Applied Biosystems, CA). The reaction volume was adjusted to 25 μL using Ultrapure distilled water (Invitrogen, CA). The reactions were performed in 8-well strips (Bio-Rad, CA) using SimpliAmp Thermocycler (Applied Biosystems, CA). Following initial denaturation step (2 minutes at 95 °C), 35 cycles of denaturation, annealing, and elongation (denaturation 30 seconds at 95 °C, annealing 30 seconds at 52 °C, elongation 30 seconds at 72 °C) were performed. A final elongation step (5 minutes at 72 °C) was performed, before being held at 4 °C. ETEC strain H10407 (O78:H11 LT+, STh+, STp+) and water was used as the positive and negative controls respectively. The amplification products were visualized by running 20 μL of amplification product in 2.5% agarose gels with ethidium bromide in 1X TBE buffer at 140 V for approximately 50 minutes and then visualizing with UV light.
ETEC positive samples were further screened using multiplex PCR for detection of colonization factors. The ETEC CFs PCR was performed as three separate panels of 5-plex PCRs. The panels are as follows: Panel 1 (CS1, CS3, CS4, CS7, CS12), Panel 2 (CS2, CS5, CS17, CS21, CFA/I), and Panel 3 (CS6, CS8, CS15, CS17/19, CS20).
We used the method described by Rodas et al. 2011 [20] with modifications. The final concentrations of the reaction mixes were 1X DreamTaq Green buffer, 0.625 U DreamTaq polymerase, 1.0 mM dNTPs (250 μM each, Denville Scientific, MA), 0.2 μM of each of five forward primers, 0.2 μM of each of five reverse primers, and 4.0 μL (approximately 100 ng) of sample DNA, brought to 25 μL final volume using Ultrapure distilled water (Invitrogen, CA). The reactions were performed in 8-well strips (Bio-Rad, CA) using SimpliAmp Thermocycler (Applied Biosystems, CA). Following initial denaturation step (2 minutes at 95 °C), 35 cycles of denaturation, annealing, and elongation (denaturation 30 seconds at 95 °C, annealing 30 seconds at 52 °C, elongation 45 seconds at 72 °C) were performed. A final elongation step (5 minutes at 72 °C) was performed, before being held at 4 °C.
Quantitative PCR
To reconfirm the PCR results and to identify any additional samples positive for CFs we rescreened all the ETEC positive samples using quantitative PCR for CFA/I, CS6, and CS21. For all qPCR reactions, PowerUp SYBR Green Master Mix (Applied Biosystems, CA) was used. The primers utilized were taken from Liu et al 2017. The final concentrations of the reaction mixes were 1X PowerUp SYBR Green Master Mix, 0.2 μM forward primer, 0.2 μM reverse primer, and 2.5 μL (approximately 10-100 ng) of sample DNA, brought to 25 μL final volume using UltraPure distilled water (Invitrogen, CA). The reactions were performed in 96-well plates (Applied Biosystems, CA) using Step One Plus Real-Time PCR System (Applied Biosystems, CA). Following initial pre-heating and denaturation steps (2 minutes at 50 °C and 2 minutes at 95 °C, respectively), 40 cycles of denaturation, annealing, and elongation (denaturation 3 seconds at 95 °C, annealing and elongation 1 minute at 60 °C) were performed. A final denaturation step (5 seconds at 95 °C) and annealing and elongation step (60 seconds at 60 °C) were performed. Finally, a melt curve was produced by increasing temperature from 60 °C to 95 °C and reading fluorescence at 0.5 °C intervals.
ETEC strain H10407 (O78:H11 LT+ STh+ STp+) was used for creation of the standard curve. H10407 was grown in LB broth overnight at 37 °C with shaking and subsequently DNA was isolated using Qiagen QIAmp DNA Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. A standard curve was created using 5-fold dilutions of the positive control DNA (5 ng/μL, 1 ng/μL, 0.2 ng/μL, 0.04ng/μL and in addition 0.00008 ng/μL). The standard curve was run on every plate in 3 replicates.
Ethical approval
Ethical revision and approval for this study was sought through the University of Zambia Biomedical Research Ethics Committee, while the National Health Research Authority provided the authorization to conduct the study. written informed consent prior to initiation of study procedures was sought from caregivers of participants.
Results
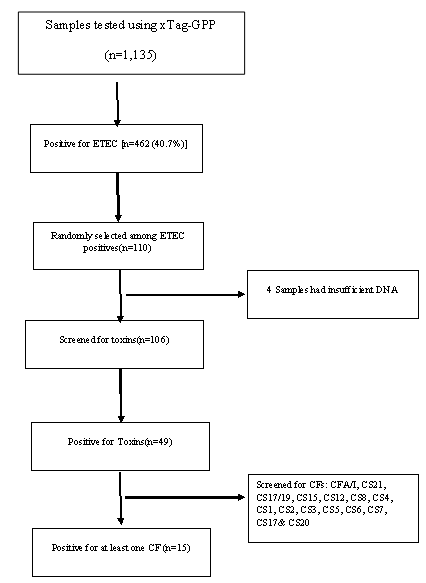
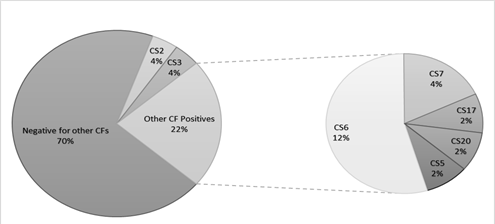
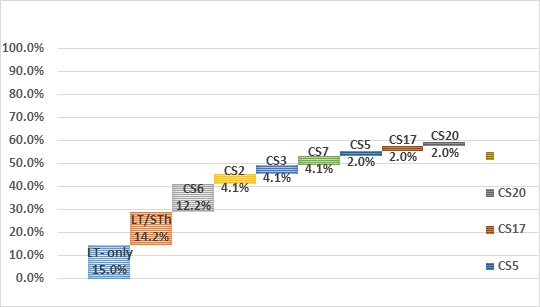
ETEC prevalence in this population was 40.7% (462 out of 1135 samples tested) using the Luminex™ xTag Gastrointestinal Pathogen Panel (see Figure 1). We randomly selected 106 samples from the samples which were positive for ETEC using Luminex™. Among these samples, 49 (46.2%) were positive for at least one of the toxins (LT/STh/STp) and/or CFs using PCR. ST was the most common toxin detected in 18(17%) with STh and STp distributed as 16 (15%) and 2 (1.9%) respectively. At least 10 samples (9.4%) had both LT and STh (See Table 2). The most common CF detected was CS6 with 6 (12.2%) positive, followed by CS2, CS3 and CS7 with 2 (4.1%) each and lastly CS5, CS17 and CS20 with one positive for each (see Figure 2). When retested using qPCR for CFA/I and CS6 the multiplex conventional PCR results matched well with the qPCR. There were two additional CS6 positives detected which were with lower copy numbers (~103 CFU/gm of stool).
The presence of CF genes differed by ETEC toxin types (Table 2). No obvious CF distribution could be noted regarding associated severity of clinical dehydration among the samples. However, five (5) out of the eight (8) CFs positive samples from children classified to be between moderate and severely dehydrated. All but one of the 8 children were one (1) year and older. CS6 was common across all subgroups (LT only, STh only and a combination of LT/STh while CS2, CS7 and CS21 where only identified in LT only and LT/STh groups respectively. Toxins and CFs were detected in children ≥12 months of age.
|
Genes |
N (%) |
|
Toxin (N=106) |
|
|
LT-only |
16 (15.1%) |
|
ST-only |
18 (17.0%) |
|
- STh |
16 (15.1%) |
|
- STp |
2 (1.9%) |
|
LT+ST |
15 (14.2%) |
|
- LT+STh |
10 (9.4%) |
|
- LT+STp |
4(3.8%) |
|
- LT+STh+STp |
1 (0.9%) |
Table 1: Summary of prevalence of toxins(n=106)
|
N |
Age(Mo) |
Sex |
LT |
STh |
STp |
CS2 |
CS3 |
CS5 |
CS6 |
CS7 |
CS17 |
CS20 |
Dehydration assessment* |
|
1 |
23 |
F |
✔ |
✔ |
✔ |
4 |
|||||||
|
2 |
15 |
M |
✔ |
✔ |
✔ |
✔ |
✔ |
2 |
|||||
|
3 |
12 |
F |
✔ |
✔ |
✔ |
3 |
|||||||
|
4 |
10 |
M |
✔ |
✔ |
3 |
||||||||
|
5 |
22 |
F |
✔ |
✔ |
2 |
||||||||
|
6 |
27 |
F |
✔ |
✔ |
✔ |
1 |
|||||||
|
7 |
18 |
F |
✔ |
✔ |
3 |
||||||||
|
8 |
25 |
F |
✔ |
✔ |
3 |
||||||||
|
Mean age (19) |
5 |
3 |
2 |
2 |
1 |
4 |
2 |
1 |
1 |
Table 2: Summary of children with at least one CF detected.
Dehydration assessment 1= Non, 2= Mild, 3=Moderate, 4= Severe
Discussion
In this study, we report for the first time the toxin and CF distributions of ETEC in the Zambian children with diarrhea. We have identified that ST is the most common toxin type expressed in this population. This is consistent with what other studies including those by Kharat et al. (2017), Platts-Mills et al. (2015) as well as a systematic review by Isidean et al. (2011) which also reported that in the GEMS sites and in Latin America and the Caribbean, the Middle East and North Africa, and South Asia, the prevalent toxin in endemic populations is ST [1, 10, 21]. Our results show that LT expressed alone or in combination with STh is expressed in ~29% of the study population, and STh alone in17% of the population. Thus, vaccine candidates that have the immunogenic LT component and multiple CFs from different classes could potentially provide a wider protection against circulating ETEC strains in this population.
CS6 was the most prevalent CFs in our population at ~12%. CS6 is an important structural gene which is critical in binding and pathophysiology of ETEC [22]. This, in our view constitutes an important vaccine target as its blockage would likely minimize the pathogen’s ability to attach to target receptors and hypothetically result in reduced pathogenic virulence. This perhaps justifies the inclusion of the CS6 targets in the constructs of the current ETEC vaccines in clinical development i.e. ACE527 and ETVax [17].
CS2, CS3 and CS7 are the CFs detected in this study population at 4.2% and is present across most ETEC subgroups (LT-only, LT/STh). Interestingly, we did not detect any CS21; this gene is reported to be the most common CF among travelers and in expected endemic pediatric diarrhea [23] as it plays a key role in bacterial self-agglutination, cell adhesions and protection from environmental stressors [24] and has been shown to play a key role in pathogenesis of ETEC diarrhea in animal models [25]. It is unclear why we could not find any of this CF in our population and as such we may only speculate: If indeed it is associated with much more severe diarrhea, it may be found among more severely ill children in tertiary admission facilities, in which case our sampled population could have missed as we recruited from primary care facilities.
Using the constructs of the two leading vaccine candidates ETVAX (CFA/I, CS3, CS5, CS6 and LCTBA (mixed with an LT B-subunit related toxoid) and the ACE527 a combination of three live-attenuated E. coli that expresses CFA/I, CS1, CS2, CS3, CS5, CS6 and the B subunit of LT, an estimated vaccine coverage of 51. 6% and 55.7% respectively (See Figure 3), with the assumption that in vaccine construct CS5 would cross protect against CS7.
Our study also reports that ETEC toxins and CFs were detected in children ≥ 12months old, (our study average age was 16.3 months (1.4 – 50.7)0 which is consistent with several studies across the globe, African continent and the region [1, 26, 27].
We also note that we got only 49 samples positive for at least one ETEC toxin using PCR and qPCR (based on the presence of LT/ST), out of the 106 total samples we tested which tested positive by Luminex™ xTag. This discrepancy could be due to the difference in the sensitivity and specificity of the two assays.
While this is the first effort to report ETEC epidemiology to this level of detail in Zambia, we recognize that our study has some methodological limitations. Firstly, the parent study recruiting participants used a passive case detection system capturing only those children who reported to health facilities with diarrhea; this means that we have no data on mild diarrhea occurring at community level without reaching the health facility. Secondly, we only targeted the classical CFs that that have been used for ETEC vaccine coverage studies, and as such we have no idea about other known CFs which were not included. Thirdly, we carried out the assays on total DNA extracted from children with MSD and had no way to account for individual strain contribution to clinical diarrhea. Thus, the toxins and CFs detected in the collected DNA could have been from different strains within the same sample. Lastly, the limitation in sample size could affect the generalizability of the results reported here, even though randomization and selection of samples was done independent of the laboratory team who tested the samples.
The study however has strengths in that we employed the use of molecular methods in detecting toxins and CFs which are more sensitive compared to phenotypic methods.
Future work
These results provide a basis for studies aimed at characterizing the ETEC isolates in Zambia using Full genome sequence analysis to identify, putative CFs especially for those isolates which would have tested negative for the known CFs.
Conclusion
Our results highlight the importance of understanding the ETEC epidemiology and strains circulating in Zambia. In addition, the prevalence of CS6 which has been isolated among isolates causing travelers’ diarrhea over a wide geographic region supports the need to do additional studies of CFs and ETEC in endemic countries and that prevalence of CS6 which has been shown to be highly conserved and important in diarrhea should be included in future multivalent ETEC vaccines.
Data availability
Data used in this study will be made available to any interested researchers upon request. To request data access, one must write to the secretary of CIDRZ Ethics and Compliance Committee, Ms. Hope Mwanyungwi (Hope.Mwanyungwi@cidrz.org) through the corresponding author (Michelo Simuyandi) mentioning the intended use for the data. The requested data should only be used for the purposes related to the original research or study. The CIDRZ Ethics and Compliance Committee will normally review all data requests within 48-72 hrs (Monday-Friday), and provide notification if access has been granted or additional project information is needed, before access can be granted.
Author contribution
MS conceptualization, data analysis draft manuscript preparation and revision
RC conceptualization, data analysis draft manuscript preparation and revision
KK draft manuscript preparation and revision
NL draft manuscript preparation and revision
CCC draft manuscript preparation and revision
JM draft manuscript preparation and revision
SBC ETEC detection, CFs testing and manuscript preparation and revision
JBV ETEC detection, CFs testing and manuscript preparation and revision
SC conceptualization, data analysis, manuscript preparation and revision
DS conceptualization, manuscript preparation and revision
Financial Disclosure
The original case-control study was funded by the Bill & Melinda Gates Foundation (Grant # OPP1033211). Detection of ETEC toxins and CFs were funded by PATH/EVI (to SC) at Johns Hopkins Bloomberg School of Public Health.
Conflict of Interest
All authors declared that they have no conflict of interest.
Acknowledgment
We are grateful to all the study participants and their caregivers. We would like to thank the CIDRZ EDVRU team for the sample preparation, storage and logistics for shipment to JHU.
References
- Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, et al. Pathogen-specific burdens of community diarrhoea in developing countries: A multisite birth cohort study (MAL-ED). Lancet Glob Heal 3 (2015): e564-e575.
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382 (2013): 209-222.
- Qadri F, Svennerholm A-M, Faruque ASG, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 18: 465–483.
- Fischer Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, et al. Global burden of childhood pneumonia and diarrhoea. Lancet 381 (2013): 1405-1416.
- Guerrant RL, DeBoer MD, Moore SR, Scharf RJ, Lima AAM. The impoverished gut--a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 10 (2013): 220.
- Investigators TMN. The MAL-ED study: A multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of. Clin Infect Dis 59 (2014): S193–S206.
- Guerrant RL. Malnutrition as an enteric infectious disease with long-term effects on child development. Nat Rev 66 (2009): 487–505.
- Fleckenstein JM. Providing Structure to Enterotoxigenic Escherichia coli Vaccine Development. J Infect Dis 216 (2017): 1–3.
- Mentzer A Von, Tobias J, Wiklund G, Nordqvist S, Aslett M, et al. Identification and characterization of the novel colonization factor CS30 based on whole genome sequencing in enterotoxigenic Escherichia coli (ETEC ). Sci Rep 7 (2017): 12514.
- Isidean SD, Riddle MS, Savarino SJ, Porter CK. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29 (2011): 6167–6178.
- Liu J, Silapong S, Jeanwattanalert P, Lertsehtakarn P, Bodhidatta L, et al. Multiplex real time PCR panels to identify fourteen colonization factors of enterotoxigenic Escherichia coli (ETEC). PLoS One 12 (2017): 1–10.
- Mentzer A von, Tobias J, Wiklund G, Nordqvist S, Aslett M, et al. Identification and characterization of the novel colonization factor CS30 based on whole genome sequencing in enterotoxigenic Escherichia coli (ETEC). Sci Rep 7 (2017): 12514.
- Fleckenstein JM, Sheikh A, Qadri F. Novel Antigens for enterotoxigenic Escherichia coli (ETEC) Vaccines. Expert Rev Vaccines 13 (2014): 631–639.
- Lundgren A, Bourgeois L, Carlin N, Clements J, Gustafsson B et al. Safety and immunogenicity of an improved oral inactivated multivalent enterotoxigenic Escherichia coli (ETEC) vaccine administered alone and together with dmLT adjuvant in a double-blind, randomized, placebo-controlled Phase I study. Vaccine 32 (2014): 7077–7084.
- Walker RI. An assessment of enterotoxigenic Escherichia coli and Shigella vaccine candidates for infants and children. Vaccine 33 (2015): 954–965.
- Leach S, Lundgren A, Carlin N, Löfstrand M, Svennerholm AM. Cross-reactivity and avidity of antibody responses induced in humans by the oral inactivated multivalent enterotoxigenic Escherichia coli (ETEC) vaccine ETVAX. Vaccine 35 (2017): 3966–3973.
- Bourgeois AL, Walker RI, Wierzba TF. Status of vaccine research and development for enterotoxigenic Escherichia coli. Vaccine 34 (2015): 2880–2886.
- Chisenga CC, Bosomprah S, Laban NM, Mwila- K, Mwaba J, Simuyandi M, Chilengi R. Aetiology of Diarrhoea in Children Under Five in Zambia Detected Using Luminex xTAG Gastrointestinal Pathogen Panel. Pediatr Infect Dis 3 (2018): 8.
- Beres LK, Tate JE, Njobvu L, Chibwe B, Rudd C, et al. A Preliminary Assessment of Rotavirus Vaccine Effectiveness in Zambia. Clin Infect Dis 62 (2016): S175–S182.
- Rodas C, Mamani R, Blanco J, Blanco JE, Wiklund G, Svennerholm AM, Sjöling Å, Iniguez V. Enterotoxins, colonization factors, serotypes and antimicrobial resistance of enterotoxigenic Escherichia coli (ETEC) strains isolated from hospitalized children with diarrhea in Bolivia. Brazilian J Infect Dis 15 (2011): 132–137.
- Kharat VB, Ahmed M, Jiang ZD, Riddle MS, Du Pont HL. Colonization factors in enterotoxigenic escherichia coli strains in travelers to Mexico, Guatemala, and India compared with children in Houston, Texas. Am J Trop Med Hyg 96 (2017): 83–87.
- Alam MM, Aktar A, Afrin S, Rahman MA, Aktar S, et al. Antigen-Specific Memory B-cell Responses to Enterotoxigenic Escherichia coli Infection in Bangladeshi Adults. PLoS Negl Trop Dis 8 (2014): 6–8.
- Guerra JA, Romero-herazo YC, Arzuza O, Gómez-duarte OG. Phenotypic and Genotypic Characterization of Enterotoxigenic Escherichia coli Clinical Isolates from Northern Colombia, South America 2014.
- Cruz-córdova A, Espinosa-mazariego K, Ochoa SA, Saldaña Z. CS21 positive multidrug-resistant ETEC clinical isolates from children with diarrhea are associated with self-aggregation , and adherence 5 (2014): 1–10.
- Guevara CP, Luiz WB, Sierra A, Cruz C, Qadri F, et al. Enterotoxigenic Escherichia coli CS21 pilus contributes to adhesion to intestinal cells and to pathogenesis under in vivo conditions. Microbiology 159 (2013): 1725–1753.
- Operario DJ, Platts-mills JA, Nadan S, Page N, Seheri M, et al. Etiology of Severe Acute Watery Diarrhea in Children in the Global Rotavirus Surveillance Network Using Quantitative Polymerase Chain Reaction. J Infect Dis 216 (2017): 220–227.
- Kotloff KL, Platts-Mills JA, Nasrin D, Roose A, Blackwelder WC, Levine MM. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine 35 (2017): 6783–6789.


 Impact Factor: * 3.1
Impact Factor: * 3.1 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 
