Genetic Variability within the ADA Gene and Left Ventricular Ejection Fraction
Article Information
Gloria-Bottini F1*, Banci M2, Neri A1, Magrini A1, Bottini E1
1Department of Biomedicine and Prevention, University of Rome Tor Vergata, School of Medicine, Rome, Italy.
2Department of Cardiology, Valmontone Hospital, Valmontone, Italy.
*Corresponding Author: Gloria-Bottini, Department of Biomedicine and Prevention, University of Rome Tor Vergata, School of Medicine, Rome, Italy
Received: 16 April 2017; Accepted: 24 April 2017; Published: 26 April 2017
View / Download Pdf Share at FacebookAbstract
Background: The role of adenosine as cardio protective factor is well established. Adenosine deaminase (ADA) contributes to the control of adenosine concentration in body fluids and, as ecto-enzyme, to the regulation of adenosine receptors activity. ADA and adenosine receptors expressions have been found down regulated in heart failure. We have studied the relationship between genetic variability within the ADA gene and left ventricular ejection fraction (LVEF).
Methods: The genotypes of three polymorphic sites (SNPs) within the ADA gene have been determined in 346 patients admitted to the hospital for cardiovascular diseases. Informed consent was obtained by the patients to participate to the study that was approved by the Council of Department of Biomedicine and Prevention. The three polymorphic sites in the ADA gene are called ADA1, ADA2 and ADA6. Each locus shows two alleles called ADA1*1 and ADA1*2, ADA2*1 and ADA2*2, ADA6*1 and ADA6*2 respectively.
Results: The joint “ADA2*2 carrier/ADA6*1/*1 genotype” shows a statistically significant lower value of LVEF as compared to other joint genotypes (p=0.004). Such association is statistically significant in subjects with coronary artery disease only.
Conclusions: The study of polymorphic sites of ADA gene could allow to detect subjects with higher risk of cardiac failure following infarction.
Keywords
ADA; LVEF; Genetic Polymorphism; CAD; Risk of Cardiac Failure
Article Details
1. Introduction
The role of adenosine as cardio protective factor is well established [1-3]. Adenosine deaminase (ADA) and adenosine kinase activities contribute to the regulation of adenosine concentration in body fluids. Adenosine binds to specific receptors: A1, A2, A3. A1 and A3 act through Gi protein and A2A and A2B through Gs protein. ADA and adenosine receptors have been found down regulated in heart failure [4].
ADA is localized on human chromosome 20q and consists of 13 exons distributed approximately on 32 Kb of DNA [5]. A number of differences among normal sequences have been found within the coding and the intronic regions of ADA gene [6]. The SNP corresponding to the presence/absence of a Taq 1 site (nt 4050-4053) in exon 1 is associated to a known functional variation and represents the basis of the common biochemical polymorphisms at ADA locus described by Spencer et al. [7]. This polymorphism is due to the presence of two common alleles ADA1*1 and ADA1*2: the corresponding phenotypes have enzymatic activity decreasing in the order: ADA11 > ADA1 21 > ADA1 2 [7]. The function of the other SNPs observed in the ADA gene has not yet been elucidated. Recent studies suggest a role of genetic variability within the ADA gene on susceptibility to coronary artery disease [8,9].
ADA is not only a cytosolic enzyme but acts as an ecto-enzyme also being present in the surface of many cell types where it regulates the extracellular concentration of adenosine and modulates the activity of receptors through interaction with other molecules [10].
We reasoned that the role of ADA in the cardiac function could be due not only to enzymatic activity i.e. the regulation of adenosine concentration, but also to ecto-enzymatic activity through interactions with adenosine receptors. Genetic variability of other sites besides the Taq 1 site in exon 1 could have an important role in this context and could influence the susceptibility to heart failure.
In the present study we have investigated three polymorphic sites (SNPs) spanning approximately 28 Kb in ADA gene in relation to the value of left ventricular ejection fraction (LVEF) an important index of cardiac function. Besides the SNP of exon 1 we have studied the Pst 1 site (ADA2) (nt 19465-19470) of intron 2 and the MluNI site (ADA6) (nt 31230-31235) of exon 6 [11].
2. Material and Methods
We have studied 346 patients admitted consecutively to the Hospital for Cardiovascular diseases (see Table 1). All subjects were from the White population of Rome. Venous blood samples were obtained after informed consent was acquired from all subjects to carry out the present study that was approved by the Council of Department of Biomedicine and Prevention . The data were collected a few years ago before the institution of an Ethical Committee. All procedures followed were in accordance with the ethical standards and with the Helsinki Declaration of 1964 and its later amendments. The number of subjects is not the same in all tables owing to random missing the data for some variables. 224 blood donors were also studied as controls.
ADA genotypes were determined by RFLP-PCR. Genomic DNA was extracted from venous blood samples collected in NaEDTA using the procedure described by Kunkel [12] with slight modifications. PCR amplification was carried out as described by Hirschhorn [11]. The details of the procedure and primers have been reported in a previous paper [13]. The alleles corresponding to the presence (+) and absence (-) of the restriction sites have been indicated as allele *1 and allele *2 respectively.
Statistical analyses were carried out by commercial software (SPSS).
3. Results
Table 1 shows clinical data of the sample study.
|
Parameters |
Proportion Mean S.D |
|
Female |
54.7% |
|
Hypertension 1 |
70.7% |
|
Diabetes mellitus |
35.2% |
|
Smoking habit |
41.7% |
|
High total cholesterol 2 |
55.7% |
|
High LDL 3 |
55.5% |
|
CAD |
59.2% |
|
Age (years) |
61.8 15.5 |
|
BMI |
27.4 5.2 |
Table 1: Clinical data in subjects with cardiovascular diseases
1 arterial tension ?130/85mmHg
2 total cholesterol > 200 mg/dl
3 LDL >130 mg/dl
Table 2 shows the values of LVEF in relation to the polymorphic ADA loci.. No significant difference in LVEF is observed within ADA1 genotypes. A lower value of LVEF is observed in carriers of ADA2*2 allele as compared to ADA2*1/*1 genotype (50.82% vs 52.79%; p=0.055). The value of LVEF is significantly lower in ADA6*2/*2 genotype than in carriers of ADA6 *1 allele ( 51.40% vs 53.60%; p=0.035).
|
|
Subjects with Cardiovascular Diseases |
Controls |
||||
|
|
Mean LVEF |
S.E. |
N° |
% |
N° |
% |
|
ADA1 locus |
|
|
|
|||
|
ADA1*1/*1 genotype |
52.10 |
0.53 |
302 |
0.88 |
193 |
0.86 |
|
Carriers of ADA1*2 allele |
53.10 |
1.34 |
40 |
0.12 |
31 |
0.14 |
|
Significance of difference |
P=0.519 |
|
|
|
||
|
ADA2 locus |
|
|
|
|||
|
ADA2*1/*1 genotype |
52.79 |
0.59 |
205 |
0.60 |
94 |
0.59 |
|
Carriers of ADA2*2 allele |
50.82 |
0.88 |
138 |
0.40 |
65 |
0.41 |
|
Significance of difference |
P=0.055 |
|
|
|
|
|
|
ADA6 locus |
|
|
|
|||
|
ADA6*2/*2 genotype |
51.40 |
0.63 |
233 |
0.67 |
101 |
0.64 |
|
Carriers of ADA6*1 allele |
53.60 |
0.80 |
113 |
0.33 |
57 |
0.36 |
|
Significance of difference |
P=0.038 |
|
|
|
||
Table 2: Left ventricular ejection fraction (%) in relation to the ADA polymorphic loci
Table 3 shows the value of LVEF in relation to the ADA2-ADA6 joint genotype. The joint genotype “ADA2*2 carrier ? ADA6*2/*2 genotype” shows a statistically significant lower value of LVEF as compared to the other joint ADA2-ADA6 genotypes.
|
ADA2-ADA6 joint genotype |
||||
|
ADA2 |
*1/*1 |
*1/*1 |
*2 carrier |
*2 carrier |
|
ADA6 |
*2/*2 |
*1 carrier |
*2/*2 |
*1 carrier |
|
|
(a) |
(b) |
(c) |
(d) |
|
|
Mean S.D |
Mean S.D |
Mean S.D |
Mean S.D |
|
|
52.40 0.71 |
54.38 1.02 |
49.42 1.21 |
52.74 1.25 |
|
Total n° |
151 |
52 |
80 |
58 |
|
Variance analysis p=0.017 |
||||
|
Post hoc LSD test |
||||
Table 3: Left ventricular ejection fraction (%) in relation to the joint ADA2-ADA6 genotype.
Table 4 shows the association of the joint of “ADA2*2 carrier-ADA6 *2/*2 genotype” with LVEF in patients with coronary artery disease (CAD) and in patients without CAD. The association is present in patients with CAD only. In subjects with CAD multivariate statistical analyses have shown that sex, age and diabetes have no effect on the association of LVEF with the joint ADA2-ADA6 genotype. The effect of ADA genotype, however, is moderate as compared to that of diabetes and age (data not shown).
|
|
“ADA2*2carriers-ADA6*2/*2 joint genotype” - |
Other ADA2-ADA6 joint genotypes - |
t-test for difference between means |
||||
|
|
Mean |
S.E |
N* |
Mean |
S.E |
N* |
|
|
All patients |
49.42 |
1.21 |
80 |
52.87 |
0.54 |
261 |
P=0.004 |
|
Patients with CAD |
45.96 |
1.52 |
52 |
49.69 |
0.72 |
150 |
P=0.015 |
|
Patients without CAD |
56.04 |
1.44 |
25 |
57.26 |
0.63 |
107 |
P=0.408 |
Table 4: The association of LVEF with the joint “ADA2*2 carrier-ADA6 *2/*2 genotype”. Comparison between patients with CAD and patients without CAD.
We have also examined the relationship of the joint “ADA2*2 carrier - ADA6*2/*2 genotype” with LVEF divided into three classes in patients with CAD (Table 5). There is high frequency of the joint genotype “ADA2*2 carrier - ADA6*2/*2 genotype” in patients with LVEF ?40 and an absence of this joint genotype in patients with LVEF > 60 (37.8% vs 0.0%, p=0.011).
|
|
|
ADA2*2 carrier |
Other joint genotypes |
|
LVEF |
40 |
17 (37.8%) |
28 (62.2% |
|
|
40-60 |
35 (24.8%) |
106 (75.2%) |
|
|
>60 |
0 (0.0%) |
16 (100%) |
|
Chi square test of independence ?2 = 9.019 df= 2 p=0.011 |
|||
Table 5: The relationship of joint “ADA2*2 carrier-ADA6*2/*2 genotype” with LVEF divided into 3 classes in patients with CAD.
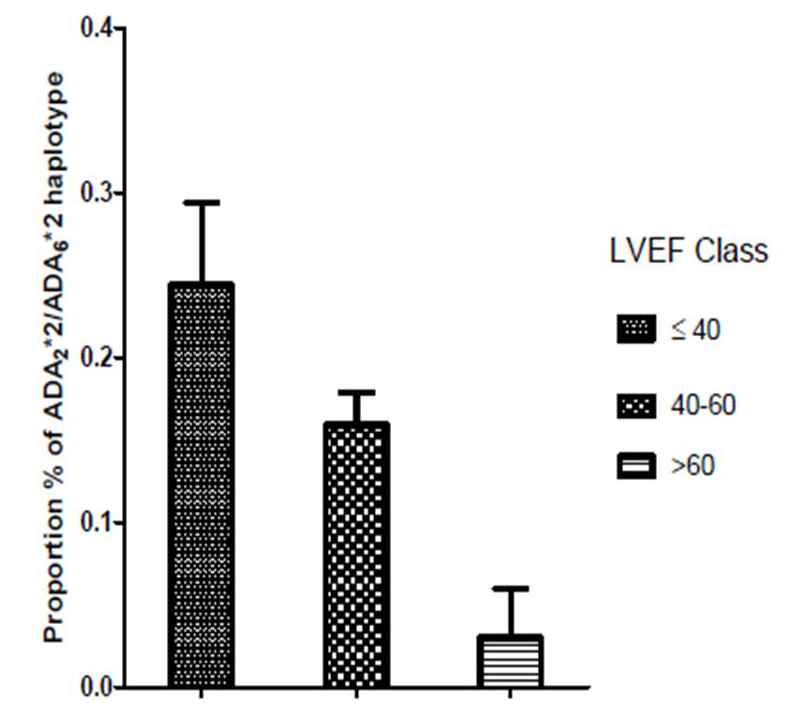
Figure 1 shows the proportion of the ADA2*2-ADA6*2 haplotype in relation to LVEF in subjects with CAD. The highest proportion of this haplotype is observed in subjects with LVEF?40 and the lowest in patients with LVEF >60.
4. Discussion
The present data suggest that in the genomic area of ADA there are sites that influence LVEF. in patients with CAD. Such association is independent by other factors associated with LVEF. No correlation with the type of treatment was observed.
We have previously observed in CAD (8) a reduced proportion of ADA2*2/ADA6*2 haplotype as compared to controls. The positive association of this haplotype with a low LVEF suggests that the reduced proportion of this haplotype is due to a lower probability of survival and not to a protection against CAD.
Since ADA2 is an intronic polymorphism and ADA6 is a synonymous substitution, these DNA modifications do not change ADA protein sequence. The association of LVEF with ADA2-ADA6 haplotype could be due to some causal site included in this area. At present, however, cannot be excluded that genetic variability in this area influences ADA enzymatic activity and in turn adenosine concentration. It is also possible that sites in this area control aminoacid sequence of the ADA protein involved in ADA properties as ecto-enzyme.
From a practical point of view, the study of polymorphic sites of ADA gene could allow to detect subjects with higher risk of cardiac failure following infarction. Further investigation of this genomic area could be rewarding.
The relatively small number of subjects examined represents the limitation of the study.
Conflict of Interest
None declared
References
- Hori M, Kitakaze M. Adenosine, the heart, and coronary circulation. Hypertension 18 (1991): 565-574.
- Kitakaze M, Hori M, Takashima S, Sato H, Inoue M, et al. Ischemic preconditioning increases adenosine release and 5'-nucleotidase activity during myocardial ischemia and reperfusion in dogs. Implications for myocardial salvage. Circulation 87 (1993): 208-215.
- Khodadadi I, Vahedi MS, Abdi M, Daneshkhah N, Rahbari R, et al. Evaluation of adenosine deaminase (ADA) isoenzymes activity and tumor necrosis factor-? (TNF?) concentration in chronic heart failure. EXCLI J 13 (2014): 58-66.
- Asakura M, Asanuma H, Kim J, et al. Impact of adenosine receptor signaling and metabolism on pathophysiology in patients with chronic heart failure. Hypertens Res 30 (2007): 781-787.
- Wiginton DA, Kaplan DJ, States JC, et al. Complete sequence and structure of the gene for human adenosine deaminase. Biochemistry 25 (1986): 8234-8244.
- Tzall S, Ellenbogen A, Eng F, Hirschhorn R. Identification and characterization of nine RFLPs at the adenosine deaminase (ADA) locus. Am J Hum Genet 44 (1989): 864-875.
- Spencer N, Hopkinson D, Harris H. Adenosine deaminase in man. Ann Hum Genet 32 (1968): 9-14.
- Saccucci P, Binczak-Kuleta A, Banci M et al. Coronary artery disease. A study of three polymorphic sites of adenosine deaminase gene. Acta Cardiol 69 (2014): 39-44.
- Gloria Bottini F, Banci M, Saccucci P, Neri A, Magrini A, et al. A study of three polymorhic sites of ADA gene in T2D subjects with coronary artery disease. J End Diab Res1 (2015): 1-3.
- Ciruela F, Saura C, Canela EI, Mallol J, Lluis C,et al. Adenosine deaminase affects ligand-induced signalling by interacting with cell surface adenosine receptors. FEBS Lett 380 (1996): 219-223.
- Hirschhorn R, Yang DR, Israni A. An Asp8Asn substitution results in the adenosine deaminase (ADA) genetic polymorphism (ADA 2 allozyme): occurrence on different chromosomal backgrounds and apparent intragenic crossover.Ann Hum Genet 58 (1994): 1-9.
- Kunkel LM, Smith KD, Boyer SH, et al.. Analysis of human Y-chromosome-specific reiterated DNA in chromosome variants. Proc Natl Acad Sci 74 (1977): 1245-1249.
- Sebastiani GD, Bottini N, Greco E, et al.. A study of Adenosine-Deaminase genetic polymorphism in rheumatoid arthritis. Int J Immunopathol Pharmacol 23 (2010): 791-795.


 Impact Factor: * 3.5
Impact Factor: * 3.5 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 14.80%
Acceptance Rate: 14.80%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks