Global DNA Methylation in Children with Complicated Urinary Tract Infection
Article Information
Kalaivani Sekar1, Sriram Krishnamurthy1*, Jharna Mandal2, Medha Rajappa3
1Department of Pediatrics, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry, India
2Department of Microbiology, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry, India
3Department of Biochemistry, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry, India
*Corresponding author: Sriram Krishnamurthy, Additional Professor, Department of Pediatrics, JIPMER, Puducherry-605006, India
Received: 19 December 2019; Accepted: 30 December 2019; Published: 02 January 2020
Citation:
Kalaivani Sekar, Sriram Krishnamurthy, Jharna Mandal, Medha Rajappa. Global DNA Methylation in Children with Complicated Urinary Tract Infection. Archives of Internal Medicine Research 2 (2019): 092-100.
View / Download Pdf Share at FacebookAbstract
Background and Objectives: The Study suggests that pathogen interceded DNA methylation changes influence the gene expression pattern contributing to disease progression. To study the association between pathogen-induced global DNA methylation variation in children with complicated Urinary tract infection and healthy children.
Methods: The study was conducted in the Department of Pediatrics of a referral hospital. For this study, 50 children as cases with complicated UTI and 50 healthy children as controls were enrolled. Whole blood genomic DNA was isolated from the cases as well as controls. Using 5-mC (methylcytosine) DNA ELISA method, the percentage of genomic DNA methylation in these children was established.
Results: Global DNA methylation level was found highly significant between cases and controls and the percentage of DNA methylation level was (Cases: 25.81 ± 6.9; Controls: 15.354 ± 3.17; P<0.001). Receiver operating curve analysis showed that the area under curve 0.92 for the total study population.
Interpretation and conclusions: The results of this study show that genomic DNA methylation varies significantly among cases and controls. This study points towards the role of DNA methylation in the pathogenesis of complicated urinary tract infection.
Keywords
Complicated urinary tract infection, DNA methylation, Epigenetic biomarker, 5-methylcytosine
Complicated urinary tract infection articles Complicated urinary tract infection Research articles Complicated urinary tract infection review articles Complicated urinary tract infection PubMed articles Complicated urinary tract infection PubMed Central articles Complicated urinary tract infection 2023 articles Complicated urinary tract infection 2024 articles Complicated urinary tract infection Scopus articles Complicated urinary tract infection impact factor journals Complicated urinary tract infection Scopus journals Complicated urinary tract infection PubMed journals Complicated urinary tract infection medical journals Complicated urinary tract infection free journals Complicated urinary tract infection best journals Complicated urinary tract infection top journals Complicated urinary tract infection free medical journals Complicated urinary tract infection famous journals Complicated urinary tract infection Google Scholar indexed journals DNA methylation articles DNA methylation Research articles DNA methylation review articles DNA methylation PubMed articles DNA methylation PubMed Central articles DNA methylation 2023 articles DNA methylation 2024 articles DNA methylation Scopus articles DNA methylation impact factor journals DNA methylation Scopus journals DNA methylation PubMed journals DNA methylation medical journals DNA methylation free journals DNA methylation best journals DNA methylation top journals DNA methylation free medical journals DNA methylation famous journals DNA methylation Google Scholar indexed journals Epigenetic biomarker articles Epigenetic biomarker Research articles Epigenetic biomarker review articles Epigenetic biomarker PubMed articles Epigenetic biomarker PubMed Central articles Epigenetic biomarker 2023 articles Epigenetic biomarker 2024 articles Epigenetic biomarker Scopus articles Epigenetic biomarker impact factor journals Epigenetic biomarker Scopus journals Epigenetic biomarker PubMed journals Epigenetic biomarker medical journals Epigenetic biomarker free journals Epigenetic biomarker best journals Epigenetic biomarker top journals Epigenetic biomarker free medical journals Epigenetic biomarker famous journals Epigenetic biomarker Google Scholar indexed journals 5-methylcytosine articles 5-methylcytosine Research articles 5-methylcytosine review articles 5-methylcytosine PubMed articles 5-methylcytosine PubMed Central articles 5-methylcytosine 2023 articles 5-methylcytosine 2024 articles 5-methylcytosine Scopus articles 5-methylcytosine impact factor journals 5-methylcytosine Scopus journals 5-methylcytosine PubMed journals 5-methylcytosine medical journals 5-methylcytosine free journals 5-methylcytosine best journals 5-methylcytosine top journals 5-methylcytosine free medical journals 5-methylcytosine famous journals 5-methylcytosine Google Scholar indexed journals Posterior Urethral valves articles Posterior Urethral valves Research articles Posterior Urethral valves review articles Posterior Urethral valves PubMed articles Posterior Urethral valves PubMed Central articles Posterior Urethral valves 2023 articles Posterior Urethral valves 2024 articles Posterior Urethral valves Scopus articles Posterior Urethral valves impact factor journals Posterior Urethral valves Scopus journals Posterior Urethral valves PubMed journals Posterior Urethral valves medical journals Posterior Urethral valves free journals Posterior Urethral valves best journals Posterior Urethral valves top journals Posterior Urethral valves free medical journals Posterior Urethral valves famous journals Posterior Urethral valves Google Scholar indexed journals bacterial infections articles bacterial infections Research articles bacterial infections review articles bacterial infections PubMed articles bacterial infections PubMed Central articles bacterial infections 2023 articles bacterial infections 2024 articles bacterial infections Scopus articles bacterial infections impact factor journals bacterial infections Scopus journals bacterial infections PubMed journals bacterial infections medical journals bacterial infections free journals bacterial infections best journals bacterial infections top journals bacterial infections free medical journals bacterial infections famous journals bacterial infections Google Scholar indexed journals kidney articles kidney Research articles kidney review articles kidney PubMed articles kidney PubMed Central articles kidney 2023 articles kidney 2024 articles kidney Scopus articles kidney impact factor journals kidney Scopus journals kidney PubMed journals kidney medical journals kidney free journals kidney best journals kidney top journals kidney free medical journals kidney famous journals kidney Google Scholar indexed journals bowel bladder dysfunction articles bowel bladder dysfunction Research articles bowel bladder dysfunction review articles bowel bladder dysfunction PubMed articles bowel bladder dysfunction PubMed Central articles bowel bladder dysfunction 2023 articles bowel bladder dysfunction 2024 articles bowel bladder dysfunction Scopus articles bowel bladder dysfunction impact factor journals bowel bladder dysfunction Scopus journals bowel bladder dysfunction PubMed journals bowel bladder dysfunction medical journals bowel bladder dysfunction free journals bowel bladder dysfunction best journals bowel bladder dysfunction top journals bowel bladder dysfunction free medical journals bowel bladder dysfunction famous journals bowel bladder dysfunction Google Scholar indexed journals Pediatrics articles Pediatrics Research articles Pediatrics review articles Pediatrics PubMed articles Pediatrics PubMed Central articles Pediatrics 2023 articles Pediatrics 2024 articles Pediatrics Scopus articles Pediatrics impact factor journals Pediatrics Scopus journals Pediatrics PubMed journals Pediatrics medical journals Pediatrics free journals Pediatrics best journals Pediatrics top journals Pediatrics free medical journals Pediatrics famous journals Pediatrics Google Scholar indexed journals Pediatric Nephrology articles Pediatric Nephrology Research articles Pediatric Nephrology review articles Pediatric Nephrology PubMed articles Pediatric Nephrology PubMed Central articles Pediatric Nephrology 2023 articles Pediatric Nephrology 2024 articles Pediatric Nephrology Scopus articles Pediatric Nephrology impact factor journals Pediatric Nephrology Scopus journals Pediatric Nephrology PubMed journals Pediatric Nephrology medical journals Pediatric Nephrology free journals Pediatric Nephrology best journals Pediatric Nephrology top journals Pediatric Nephrology free medical journals Pediatric Nephrology famous journals Pediatric Nephrology Google Scholar indexed journals
Article Details
Abbreviations:
CAKUT- Congenital anomalies of kidney and urinary tract; HDN- Hydronephrosis; 5-mC- methylcytosine; PUV- Posterior Urethral valves; ROC- Receiver operating characteristics; UTIs- Urinary; VUR- Vesicoureteral reflux
1. Introduction
Urinary tract infections (UTIs) are one of the most common and severe bacterial infections in children and account for significant morbidity. Uropathogenic E. coli (UPEC) are the most documented uropathogen in urinary tract infections [1]. UTIs are an important cause of fever in children. They might reflect an underlying structural defect in the kidney and urinary tract or bowel bladder dysfunction [2]. Diagnosis is the crucial step in the treatment of UTI, infants and children with developing immune systems are prone to recurrent UTI. The pathogenesis of UTI involves the interaction between the virulence factors and host defense mechanisms through which the immune response to UTI is initiated [3]. Despite various defense mechanisms against uropathogen, the infection can continue within the urinary tract and can lead to recurrent infections. As a consequence, a substantial proportion of children are unable to create protective immunity for uropathogens, and this inability to initiate an appropriate adaptive immune response may be due to the host exposure to infection [1, 4]. Although virulence of the pathogenic bacteria and underling structural anomalies are considered important factors in the pathogenesis of UTI, it is being increasingly recognized that DNA methylation could play a vital role [3]. DNA methylation is one of the epigenetic modifications and involves the addition of the methyl group to cytosine or adenine nucleotide. This does not alter the genetic backup i.e., a change in phenotype without a change in genotype. This can be influenced by environmental factors importantly stress, food habits, age and infection conditions or unknown reasons [5, 6]. In recent years research has identified that the host response to infection is a cause for infection-induced host DNA methylation [7]. Such infection-induced modifications are seen in Helicobacter pylori infection of the stomach, Schistosoma infection of the bladder and UPEC infection of uroepithelial cells [8, 9, 10]. DNA 5-methylcytosine (5 mC) modifications are being studied for their pathogenesis in complex disorders [11]. Recent studies have shown that CALCA (Calcitonin related polypeptide alpha) gene promoter methylation varied with different types of bacterial sepsis [12]. Another study has evaluated the utility of DNA methylation in neonatal sepsis [13]. This alteration in DNA methylation level can apply for the development of biomarker and also for disease prognosis. DNA methylation in response to UTI infection is still at its formative stage and no DNA methylation-based biomarker for UTI exists [14]. New genetic markers like single nucleotide polymorphism (SNP) genotyping and DNA methylation are extensively studied for specificity, sensitivity, and prognostic efficacy. The study aimed to investigate the global DNA methylation level by ELISA method in children with complicated UTI in comparison with healthy children.
2. Materials and Methods
The study was conducted in the Department of Pediatrics of a tertiary care referral hospital. The study was approved by the Scientific Advisory Committee and Institute Human Ethical Committees. Children aged 1m–18 years (either gender) with clinical symptoms consistent with complicated UTI (as defined by the revised guidelines formulated by the Indian Society of Pediatric Nephrology) were recruited as cases [15]. Children who had received steroids/immunosuppressant in the last 90 days or presenting 48 hours after initiation of antibiotic therapy or chronic kidney disease stage 4 and 5 were excluded. The inclusion criteria for healthy children (1m–18 years, either gender) without the signs of complicated UTI and other infections, not having structural anomalies of the kidney and urinary tract (Figure 1).
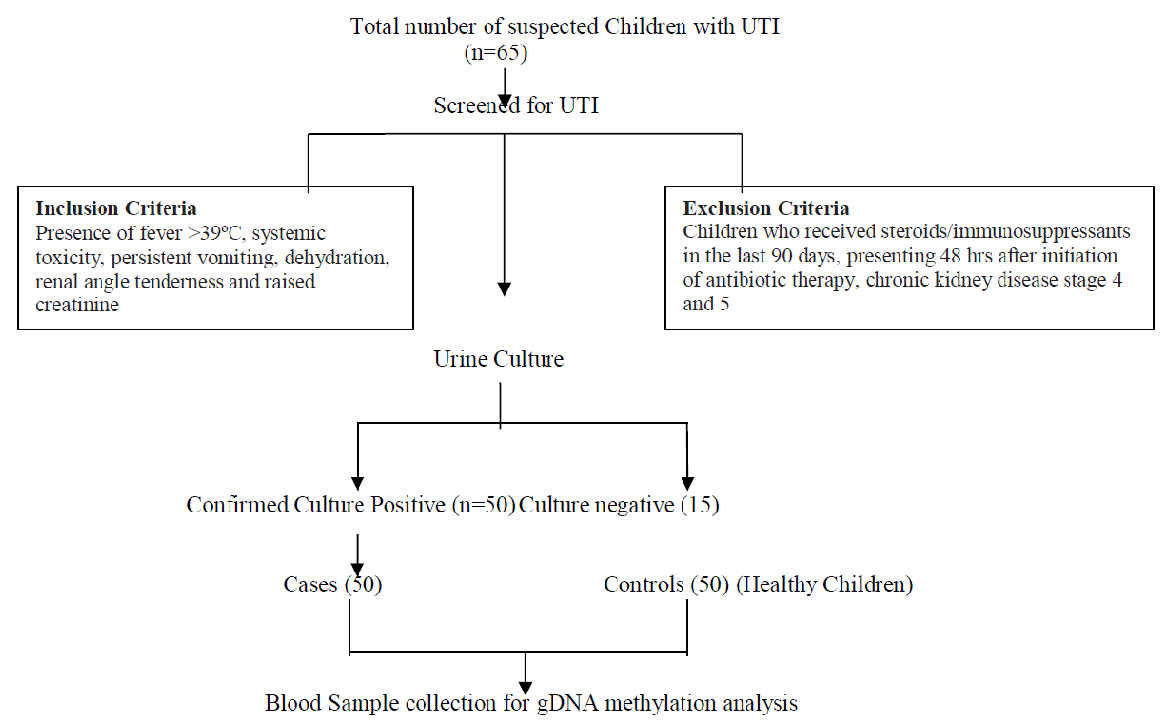
Figure 1: Flow chart depicting inflow of subjects into the study.
Fifty children as cases with complicated UTI and fifty children as controls were enrolled in this study. After obtaining informed signed consent from the parents and assent from the children, 1.5mL of the blood sample was collected in an EDTA vacutainer from the cases (complicated UTI) as well as controls. DNA was isolated from whole blood (300μL) using Genomic DNA Mini kit (Favorgen Biotech corp., Taiwan) as per the manufacturer’s protocol. The genomic DNA concentration was measured using Nanodrop 2000 spectrophotometer (Thermo Scientific, USA) and was stored at –40°C until analysis. We detected the concentration of 5-methylcytosine level in both cases and controls by 5-mC DNA ELISA kit (Zymo Research, USA) according to the manufacturer’s instructions. The 5?mC level in DNA of all samples was reported as the number of methylated cytosines relative to the cytosine genomic content (percent). The negative (unmethylated) and positive controls (methylated) consisted of 100ng/μL of double-stranded DNA. All samples, including methylated and unmethylated standards (used to generate a standard curve for the quantification of the unknown samples), was prepared by mixing negative and positive controls at different proportions. The input DNA concentrations for the 5?mC assays were 100ng of DNA which was added to separate PCR tubes and the final volume was made to 100μL with 5-mC coating buffer. All samples were added using the same amount of DNA. The DNA samples were denatured by heating at 98°C for 5 min in a thermal cycler (Agilent Surecycler 8800, CA, USA). After denaturation, the samples were transferred immediately to the ice for 10 min. Then the entire denatured DNA was added to the 96-well microtiter plate, covered with foil and incubated at 37°C for 1h. The contents of the wells were discarded after the 1h of incubation period and the wells were washed three times with 200μL of 5-mC ELISA buffer. Thereafter 200μL of 5-mC ELISA buffer was added to the wells and the plate was covered, incubated at 37°C for 30 min. After incubation, the buffer was discarded from the wells and 100μL of the antibody mix (Anti- 5-Methylcytosine and secondary antibody in 5-mC ELISA buffer in the ratio of 1:2:2000) was added to each well and incubated at 37°C for 1h. After incubation, the antibody mix from the wells was discarded and washed three times with 200μL of 5-mC ELISA buffer. The absorbance values were measured at wavelengths (405–450nm) after the addition of the 100μL HRP developer kept at room temperature for 10–60 min for color development using spectrophotometer (Thermo Fisher Scientific Inc.). The results were measured in triplicate and the average was taken. All statistical analysis was performed in SPSS v20 and MS Excel at a 95% confidence interval at a 5% level of significance. Categorical data were expressed as numbers and percentages. To estimate the significant differences in DNA methylation level between cases and controls, Independent samples‘t’test was carried out. Percentage of genomic DNA methylation level was expressed in mean with a standard deviation between cases and controls. Paired t-test was used to evaluate the significant difference in %5-mC within the cases. Receiver operating characteristics (ROC) curve analysis was used to evaluate the cut-off value of %5-mC to distinguish between cases and controls.
3. Results
The children enrolled in this study concerning frequency and percentage-wise distribution of demographic variables among children with complicated urinary tract infection and healthy controls were depicted in (Table 1). Regarding age 42 out of 50 (84%) in cases and 41 out of 50 (82%) in controls belong to the age group less than 5 years and (16%) in cases and (18%) in controls belong to the age group greater than 5 years. The mean age group of cases is 33.06months (2.75years) and 38.98 months (3.24years) in controls.
|
Sl. No. |
Variables |
Cases |
Controls |
p-value |
|||
|
n |
% |
n |
% |
||||
|
1 |
Age |
||||||
|
Less than 60 months (<5 years) |
42 |
84 |
41 |
82 |
0.790 |
||
|
More than 60 months (>5 years) |
8 |
16 |
9 |
18 |
|||
|
Mean Age In Years |
33.06 months |
38.98 months |
0.439 |
||||
|
2 |
Gender |
||||||
|
Male |
31 |
62 |
30 |
60 |
0.838 |
||
|
Female |
19 |
38 |
20 |
40 |
|||
|
3 |
Urine Culture Positive |
||||||
|
E.coli Klebsiella pneumoniae Enterococcus Enterobacter Citrobacter koseri |
37 6 4 2 1 |
74 12 8 4 2 |
– |
– |
– |
||
|
4 |
Types of UTI |
||||||
|
a) Complicated UTI b) Recurrent UTI c) Breakthrough UTI |
41 7 2 |
82 14 4 |
– |
– |
– |
||
|
5 |
CAKUT |
||||||
|
VUR Non-obstructive HDN PUV |
10 10 3 |
20 20 6 |
– |
– |
– |
||
|
6 |
Renal Scars |
6 |
12 |
– |
– |
– |
|
CAKUT-Congenital anomalies of kidney and urinary tract; VUR-Vesicoureteral reflux; HDN-Hydronephrosis; PUV-Posterior Urethral valves
Table 1: Demographic characteristics of enrolled children.
Among 65 cases with clinical suspicion of complicated UTI, Urine culture was positive in 50 cases. Thirty-seven of them had E.coli (74%), and remaining were Klebsiella pneumoniae (12%), Enterococcus (8%), Enterobacter (4%), Citrobacter koseri (2%). The comparison of genomic DNA methylation level was assessed between the cases and controls were shown in Table 2.
|
Parameters |
% of DNA methylated (Mean±SD) |
P value |
|
Cases (n=50) |
25.81 ± 6.92 |
<0.001 |
|
Controls (n=50) |
15.35 ± 3.17 |
Table 2: Comparison of percentage of DNA methylation level between cases and controls.
The mean and standard deviation of the DNA methylation level between cases and controls was (25.81 ± 6.92) and (15.35 ± 3.17) the independent samples ‘t’ test shows the results, p-value (<0.001) are significant (Figure 2).
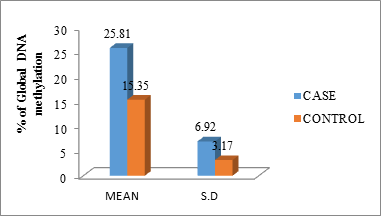
Figure 2: Percentage of global DNA methylation of cases and controls in mean with standard deviation.
The genomic DNA methylation level was compared with complicated UTI and its outcome and expressed in median with inter quartile range (IQR). The genomic DNA methylation level was assessed among cases with the severity of UTI with such as CAKUT, UPEC and presence of renal scar. No significant difference in percentage of global DNA methylation level was found within cases included in this study in relation with clinical outcome (Table 3).
|
Parameters |
% of DNA methylation Median with IQR |
P value |
|
CAKUT (23) Without CAKUT (27) |
24.5(20.8–31.1) 24.7(20.8–28.5) |
0.704 |
|
UPEC causing UTI (37) Other organisms causing UTI (13) |
25.3(22.7–31.08) 23.1(18.7–28.6) |
0.838 |
|
Renal Scar (6) Without renal Scar (44) |
25.4(22.3–29.2) 24.7(20.12–31.00) |
0.384 |
Table 3: Comparison of percentage of DNA methylation level with complicated UTI and its outcome.
The Frequency and percentage-wise distribution of the level of DNA methylation between cases and controls were shown in Table 4. In cases, 27 (54%) had DNA methylation level of 21–30% whereas in control 44 (88%) had a 10–20% level of DNA methylation. Less than 10% of DNA methylation was not seen in cases. In controls, the complete absence of genomic DNA methylation level was seen in 31–40% and >40%.
|
% of DNA Methylation |
Cases |
Controls |
P-Value |
||
|
|
n |
% |
n |
% |
<0.001 |
|
<10 |
0 |
0 |
2 |
4 |
|
|
10–20 |
10 |
20 |
44 |
88 |
|
|
21–30 |
27 |
54 |
4 |
8 |
|
|
31–40 |
12 |
24 |
0 |
0 |
|
|
>40 |
1 |
2 |
0 |
0 |
|
Table 4: Frequency and percentage-wise distribution of DNA Methylation between cases and controls.
Receiver Operating Curve (ROC) was generated to examine the precision of the 5 mC test. ROC analysis showed a cut-off value of 15.56% genomic DNA methylation between cases and controls. It can be used as a diagnostic threshold value to distinguish cases from controls, with 92% sensitivity and 40% specificity (P<0.001) (Figure 3).
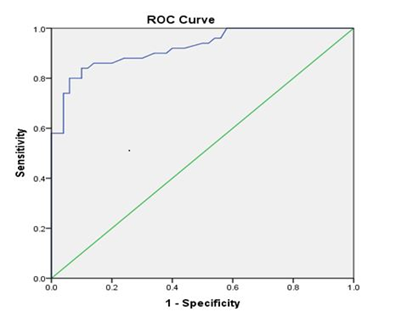
Figure 3: Receiver operating curve shows a cut off value to distinguish the percentage of DNA methylation between cases and controls (cut off value = 15.56 % with 92% sensitivity and 40% specificity, P<0.001). The area under the curve was 0.92.
4. Discussion and Conclusion
Methylation at the 5´ position of cytosine in DNA is a relatively stable epigenetic modification that plays a role in regulating gene expression, development and disease processes. In the case of cancer, DNA methylation is being established as a potential diagnostic marker [16, 17]. In human, most of the CpG sites and CpG islands in the genome are unmethylated, and the genes associated with it are found to be expressed in the normal individual. In contrast, discrete CpG sites are found to be mostly methylated, affecting gene expression. Global hypomethylation and gene-specific hypomethylation may lead to over-expression of genes that contribute to disease progression. The global hyper? and hypomethylation equation is a decisive checkpoint for the host to activate or suppress the appropriate immune response against infection. The extent of DNA methylation determines the level of gene expression, higher the DNA methylated, lower its expression. The 5-mC DNA proportion is directly comparable to the percentage of methylated DNA [18, 19, 20]. The present study was an attempt to elucidate the percentage of genomic DNA methylation in children with complicated UTI in comparison with healthy controls. Children with complicated UTI and healthy children were enrolled in the study and their genomic DNA was analyzed for the concentration of 5-mC. The cases were highly methylated when compared with controls and the level of DNA methylation is distinguishable between cases and controls. The DNA methylation level found in this study might have been pathogen-induced as a result of the host response to infection. The variation in the level of genomic DNA methylation found in this study among cases and controls states that children with UTI are significantly methylated leading to variation in host immune behavior via underlying gene expression or it may influence immune genes to respond against UTI in children. ROC was generated to scrutinize the accuracy of the 5 mC quantification. A cut off value of 15.56% for gDNA methylation was found to distinguish cases from controls with 92% sensitivity and 40% specificity with a statistically significant difference. The 5-mC quantification test performed in this study can differentiate cases from controls effectively. Here we investigated the extensively studied epigenetic modifications, namely 5-mC, on a global scale. This is the first study to evaluate DNA methylation in complicated UTI and offers useful insights into the pathogenesis of UTI. Our study has some limitations. DNA methylation in UTI is a novel topic of research and has not been described earlier in literature. Hence, we chose a convenience sample size. For logistic reasons, we studied the percentage of DNA methylation in blood samples only (and not in shed uroepithelial cells in urine). Moreover, children with complicated UTI often have signs of systemic toxicity which makes the study of blood samples a reasonable approach to assess global DNA methylation in blood. Further research on the role of DNA methylation and UTI should focus on identifying and targeting specific CpG sites methylation to improve the efficiency of DNA methylation as an epigenetic biomarker for UTI. These findings require confirmation in future studies by evaluating DNA methylation at gene loci specific by using bisulfite sequencing or pyrosequencing could provide a better strategy to decipher the secrets behind epigenome and its role in UTI.
Acknowledgment
This study was supported by an intramural grant from JIPMER which is gratefully acknowledged.
Conflicts of Interest
No Conflict of Interest
References
- Spencer JD, Schwaderer AL, Becknell B, Watson J, Hains DS. The innate immune response during urinary tract infection and pyelonephritis. Pediatric Nephrology 29 (2014): 1139-1149.
- Chang SL, Shortliffe LD. Pediatric urinary tract infections. Pediatric Clinics of North America 53 (2006): 379-400.
- Bien J, Sokolova O, Bozko P. Role of uropathogenic Escherichia coli virulence factors in development of urinary tract infection and kidney damage. International Journal Nephrology (2012): 1-15.
- Hannan TJ, Mysorekar IU, Hung CS, Isaacson-Schmid ML, Hultgren SJ. Early severe inflammatory responses to uropathogenic E. coli predispose to chronic and recurrent urinary tract infection. PLoS Pathogens 6 (2010): e1001042.
- Tollefsbol T. Handbook of epigenetics: the new molecular and medical genetics. Oxford, UK: Academic Press, Elsevier Inc (2011).
- Simmons D. Epigenetic in?uence and disease. Nature Education 1 (2008): 6.
- Stein RA. Epigenetics – the link between infectious diseases and cancer. Journal of American Medical Association 305 (2011): 1484-1485.
- Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Research 70 (2010): 1430-1440.
- Zhong X, Isharwal S, Naples JM, Shiff C, Veltri RW, Shao C, et al. Hypermethylation of Genes detected in urine from Ghanaian adults with bladder pathology associated with Schistosoma haematobium infection. PLoS ONE 8 (2013): e59089.
- Tolg C, Sabha N, Cortese R, Panchal T, Ahsan A, Soliman A, et al. Uropathogenic E.coli infection provokes epigenetic downregulation of CDKN2A (p16INK4A) in uroepithelial cells. Laboratory Investigation 91 (2011): 825-836.
- Feinberg AP. Genome-scale approaches to the epigenetics of common human disease. Virchows Archiv 456 (2010): 13-21.
- Tendl KA, Schulz SM, Mechtler TP, Bohn A, Metz T, Greber-Platzer S, et al. DNA methylation pattern of CALCA in preterm neonates with bacterial sepsis as a putative epigenetic biomarker. Epigenetics 8 (2013): 1261-1267.
- Dhas BB, Antony HA, Bhat V, Newton B, Parija SC. Global DNA methylation in neonatal sepsis. Indian Journal of Pediatrics 82 (2015): 340-344.
- Tolg C, Bagli DJ. Uropathogenic E.coli infection: potential importance of epigenetics. Epigenomics 4 (2012): 229-235.
- Indian Society of Pediatric Nephrology, Vijayakumar M, Kanitkar M, Nammalwar BR, Bagga A. Revised statement on management of urinary tract infections. Indian Pediatrics 48 (2011): 709-717.
- Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harbor Perspectives in Medicine 2 (2012): a010272
- Dehan P, Kustermans G, Guenin S, Horion J, Boniver J, Delvenne P. DNA methylation and cancer diagnosis: new methods and applications. Expert Review of Molecular Diagnostics 9 (2009): 651-657.
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim et Biophysica Acta 1775 (2007): 138-162.
- Scharer CD, Barwick BG, Youngblood BA, Ahmed R, Boss JM. Global DNA methylation remodeling accompanies CD8 T cell effector function. Journal of Immunology 191 (2013): 3419-3429.
- Philips T. The role of methylation in gene expression. Nature Education 1 (2008): 116.


 Impact Factor: * 3.6
Impact Factor: * 3.6 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 