Inference from Open-Source Sequence Data on the Genetic Epidemiology of COVID-19 Infection in Africa
Article Information
Chigozie J Nwachukwu1*, Raina McIntyre2, Haley Stone2
1Achieving Health Nigeria Initiative, Awka, Anambra State, Nigeria
2BioSecurity Research Programme, Kirby Institute, University of New South Wales, Sydney, Australia
*Corresponding Author: Chigozie J Nwachukwu, Achieving Health Nigeria Initiative, Awka, Anambra State, Nigeria.
Received: 06 February 2023; Accepted: 13 March 2023; Published: 29 March 2023
Citation: Chigozie J Nwachukwu, Raina McIntyre, Haley Stone.Inference from Open-Source Sequence Data on the Genetic Epidemiology of COVID-19 Infection in Africa. International Journal of Plant, Animal and Environmental Sciences 13 (2023): 13-22.
View / Download Pdf Share at FacebookAbstract
Between late December 2019 when it was first reported, to early September 2020, severe acute respiratory syndrome coronavirus-2 (SARSCoV- 2), the causative agent of coronavirus disease-2019 (COVID-19), reportedly infected over 10 million people globally. In Africa, more than 300,000 infections occurred within the period, from which several genetic sequences of the virus were generated. During disease outbreak, phylogenetic reconstruction of genomic data can assist in making epidemiological inferences about time of pathogen introduction, epidemic growth rate and temporal-spatial spread of the infection. In this work, we studied the genetic epidemiology of COVID-19 in Africa. Genetic sequence data of SARS-CoV-2 and metadata from African countries were obtained from publicly accessible sequence database hosted by the GISAID initiative. Whole genome sequences were subjected to multiple sequence alignment and the aligned sequences used to construct a Maximum Likelihood phylogenetic tree based on the general time reversible model. Of the 227 genetic sequences obtained for 9 African countries (DRC=133, Senegal=23, South Africa=20, Ghana=15, Tunisia=6, Algeria=3, Gambia=3, Egypt=2 and Nigeria=2), 220 were whole genome sequences while 7 were partial genome sequences of the surface glycoprotein S. Majority of the viruses (58.1%) belonged to the G clade of GISAID classification. Phylogenetic analysis confirmed multiple introductions of the virus to the continent from multiple external sources prior to local adaptation and spread. The very close alignment of three viruses - Ghana/1659_S14/2020|EPI_ISL_422405, DRC/KN0054/2020|EPI_ ISL_417437, and South_Africa/R05475/2020|EPI_ISL_435059 – to the reference Wuhan strain on the time tree, suggests possible introduction and circulation of the virus into the continent much earlier than when the first case in the continent was announced in February 15 2020. In conclusion, this study provided evidence to support multiple introductions of SARSCoV- 2 into Africa, and further suggests that the virus may have already been circulating in the continent prior to official reporting of the first case. Also, there is strong impression to infer likely genetic adaptation of the virus in the continent that may account for the close clustering of isolates from different countries of the continent.
Keywords
Africa; SARS-CoV-2; Phylogenetics; Viral transmission; Spread
Africa articles; SARS-CoV-2 articles; Phylogenetics articles; Viral transmission articles; Spread articles
Article Details
1. Introduction
In late December 2019, the World Health Organisation (WHO) was notified of cluster of cases of pneumonia of unknown aetiology in the Hubei province of China [1]. They were later confirmed to be caused by a novel coronavirus, severe acute respiratory syndrome coronaviruses-2 (SARS-CoV-2) [2] and the associated disease named coronavirus disease-2019 [COVID-19] [3]. Up till early September 2020, over 10 million infections have been reported globally [4]. The first case of COVID-19 in Africa was reported on 15 February 2020 in Egypt, and as of ending of June 2020, Africa has recorded over 300,000 cases [4]. As of early September 2020, a total of 6,155 COVID-19 associated deaths have been recorded in Africa with over 150,000 recoveries. With a global Case-Fatality Ratio (CFR) of 5.0%, Africa’s contribution to worldwide COVID-19 associated death of 503,862 stood at 1.2%.
In the midst of efforts to contain the pandemic, researchers worked assiduously to understand various characteristics of the virus including its pathogenicity, immunogenicity, pathobiology, and epidemiological characteristics. Early studies on the clinicoepidemiological characteristics of the virus reported mean incubation period of 5.2 days, ranging up to 12.5 days in 95th percentile of the distribution [5]. This finding formed the basis of the recommendation of 14 days isolation period for exposed persons.
In terms of geographical spread, COVID-19 was reported from 55 African countries and territories (including Mayotte), with South Africa having the highest number of cases in sub-Sahara Africa with total confirmed cases of over 350,000 and 4,948 deaths as of end of June 2020 [6]. According to the Africa Centre for Disease Control and Prevention (Africa CDC), eight countries in Africa are reporting case fatality rate that is similar or higher than the 4.2% obtained globally. These include: Chad (8.4%), Liberia (6.3%), Sudan (6.3%), Niger (6.2%), Burkina Faso (5.0%), Egypt (4.9%), Mali (4.9%) and Algeria (4.7%) [7]. Contrary to situation seen in past epidemics, the continent demonstrated an improved laboratory diagnostic capacity in response to COVID-19, with ability for local viral sequencing capacity in several countries. This was an improvement from situations in years past whereby external collaboration was usually needed for definitive diagnosis of a disease outbreak [8]. One outcome of this situation was the array of SARS-CoV-2 partial and whole-genome sequence data that was submitted to the public database of the GISAID Initiative (formerly Global Initiative on Sharing All Influenza Data) [9] by scientists from various laboratories in Africa. Analysing these sequence data is expected to furnish information on the genetic epidemiology of the SARS-CoV-2 infection in Africa.
Molecular or genetic epidemiology is concerned with how genomic, genetic, and other molecular attributes contribute to the aetiology of a disease, its distribution and approach to prevention [10]. Through the analysis of molecular data, inferences could be made on the rate of evolution of a viral pathogen, the evolutionary changes influencing host and tissue specificity, and the pattern of transmission within human population. An important tool in achieving this is phylogenetic studies. Phylogenetic analysis is a critical tool for understanding the historical evolution of viruses and serves as the basic building block for their classification [2,10].
To inform public health intervention, genome sequence data requires rapid analyses and wider dissemination of the results obtained. Since the identification of the aetiological agent of COVID-19 and curation of sequence data on GISAID, large scale phylogenetic analyses of the sequences have been undertaken by NextStrain [11] an open-source project aimed at harnessing “the scientific and public health potential of pathogen genome data” [12]. Inferences from the analyses showed that, firstly, outbreaks in far-flung geographical locations are intertwined. Secondly, there were multiple introductions of the virus to communities through migration and human travel. Thirdly, not all introduced variants resulted in local spread while few others spawn local transmission. And lastly, once local transmission is established, these send off their own sparks into other communities [13]. Thus, the interest of the current work is to understand how outbreaks in Africa which were seeded from other continents, especially Europe [8,14], have interacted to produce the predominant pattern in Africa then. Doing so will inform robust surveillance system that could alert to genetic changes or trans-continental introduction of new variants.
Relying on two main sources of publicly available data, our objective was to compare the genetic epidemiology of COVID-19 infection in African countries through phylogenetic studies. Our result showed that the data curated by the COVID-19 Data Working Group [15] was inadequate for making inferences on the general descriptive epidemiology of COVID-19 in Africa on one hand. On the other hand, with exception to wide disparities in the numbers of SARS-CoV-2 sequences submitted from few African countries to the GISAID database [16], the quality of the data enabled inferential deductions that may reflect the epidemiological pattern of the COVID-19 pandemic in Africa.
2. Materials and Methods
2.1 Source of data and data retrieval
Data for this study were obtained from two main sources – GISAID database [16] and Open COVID-19 Data Working Group Repository [15] (detailed acknowledgment available in Table S1), supplemented by COVID-19 infection data from WHO-OCHA [17] and population data from the World Bank’s Health Nutrition and Population Statistics database [18]. The GISAID database is a publicly available open-access platform for submission of genome sequence data. The COVID-19 epidemiological line list is a centralised repository of individual-level information on patients with laboratory-confirmed COVID-19. The data are openly available [15], and a live version of the data record, which is continually updated, can be downloaded from the group’s github repository.
On 27 May 2020, the GISAID database was accessed and the EpiCoV tab selected. On the browser pane, Africa was typed in to filter the data to display only entries from Africa, excluding contents from other parts of the world. The 227 sequences from Africa as of that date were downloaded manually as individual files in Fasta format. The downloaded files were then imported into Geneious prime® (Geneious version 2020.2 created by Biomatters. Available from https://www.geneious.com) for editing. Also, the reference strain, China/WHU01/2020/EPI_ISL_406716 was downloaded separately and imported into the Geneious® software.
Similarly, latest data up to 17 July 2020 on global COVID-19 epidemiological line list was downloaded from the github repository of the COVID-19 Data Working Group. This contained data up till end of May 2020. These were filtered to select only for African countries and data for African countries were extracted into a separate Excel sheet for analysis.
2.2 Epidemiological analysis on line-list data
The data on the line list was analysed for count of cases and number of cases per country. However, age and gender distribution could not be meaningfully estimated because these data points are not available in several of the line list entries. Data from this source were used to plot the number of daily cases in Africa up to the end of May 2020 and the number of cases per country. The results were compared to WHO-OCHA daily cases report from African countries for the period up to April 30, 2020.
2.3 Updating and analysing sequence metadata
The manually downloaded sequence data contained only the nucleotide bases without metadata. For each sequence, metadata including age, sex, location, and clades were updated in Geneious prime® (Geneious version 2020.2 created by Biomatters. Available from https://www.geneious.com) and on Excel file by triangulating back to the GISAID database. The age and sex distribution of the sequence data were analysed and the count of lineages and clades from each country annotated.
2.4 Sequence alignment
Sequences were manually filtered and partial gene sequences with sequence size of 1kb were excluded. Whole genome sequences with average sequence length of 29,000 to 30,000 were selected and aligned with the MUSCLE algorithm [19] in MEGA version X software [20] using default settings.
2.5 Building of phylogenetic tree
In MEGA version X software [20], the aligned sequences were used to estimate the tree by the maximum likelihood (ML) model in which gaps and missing data were set for complete deletion and Neighbour-joining (NJ) tree specified as preferred tree to use. The General Time Reversible (GTR) model [21] was used to fit the tree. By applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances obtained from the Maximum Composite Likelihood (MCL) approach, initial tree(s) for heuristic search were obtained automatically and the topology with superior log likelihood value was selected. Evolutionary rate differences among sites [5 categories (+G parameter=200.0)] was modelled by discrete Gamma distribution. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site and the tree with the highest log likehood (-586456.26) was displayed.
2.6 Tree visualisation and editing
The phylogenetic tree obtained from MEGA version X [20] was exported in Newick tree format into FigTree version 1.4.4 [22] where it was edited and time scale added to infer the diversity time based on the length of the tree.
3. Results
3.1 Cumulative number of daily cases of COVID-19 in Africa up to end of May 2020 and number of cases per country
Based on the data obtained from the Open COVID-19 Data Working Group, we estimated the number of daily cases of COVID-19 in Africa up to the period (end of May 2020) that the data obtained on 17 July 2020 was curated. The number of cases per country was also estimated from the data. This was compared to the number of daily cases collated by the WHO-OCHA [17] based on data downloaded on 19 July 2020 that were updated up to end of May 2020.
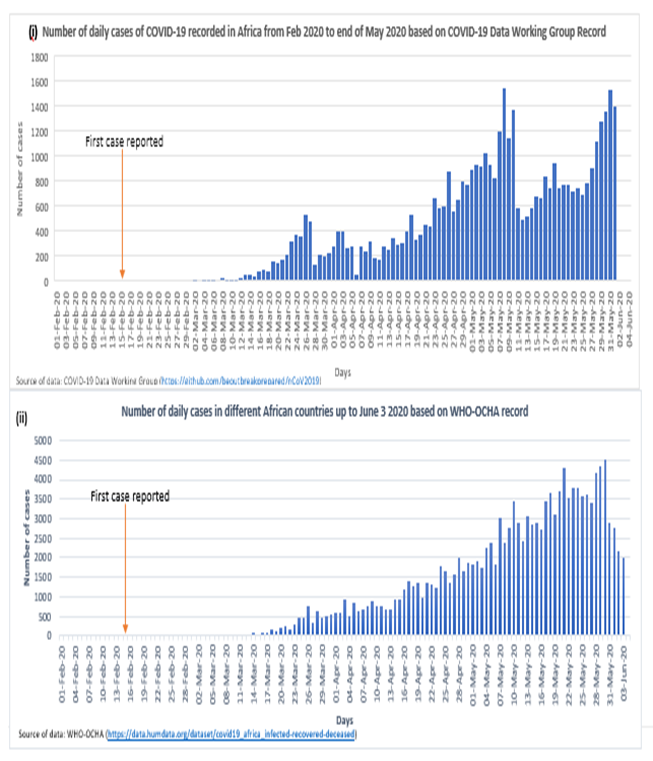
Table 1 shows the cumulative number of cases per country based on the data from the earlier mentioned sources while the number of daily cases is presented in Figure 1 below. The total number of cases of COVID-19 in Africa as of July 3, 2020 is 142,245 based on the WHO-OCHA data. The top 5 countries with highest number of cases are South Africa, Egypt, Nigeria, Algeria, Ghana, and Morocco with 20.7%, 20.1%, 7.9%, 6.8% and 6.0% of all the cases in the continent, respectively. However, estimation of infection rate show that rate of infection is highest in the horn of Africa country of Djibouti, with an infection rate of 388.2 per 100,000 population (see Table 1 and Figure 1). There was gross under-reporting of number of cases in the data curated by the COVID-19 Data Working Group when compared to the WHO-OCHA data. Apart from lack of data from some countries, few numbers were also reported for many countries when compared to the WHO-OCHA data. This may be due to the fact that the curated data were often from official and non-official sources [15], and may be limited by internet access at the different countries.
From when the first case in Africa was reported on February 15, number of daily cases has progressively increased reaching a record level of over 4500 cases in one day on May 30 as reflected from the WHO-OCHA data [see Figure 1.ii]. When viewed from the Open COVID-19 Data Working Group data, the number of daily cases appears to have reduced in the second week of May before picking up again [Figure 1 (i)]. This however was not the case as the WHO-OCHA data show unrelenting increase in the number of cases.
|
Country (Population‡) |
Number of cases from COVID-19 Data working group |
Number of cases from WHO-OCHA |
Percentage of total cases (%)† |
Rate of infection per 100,000 population |
|
South Africa (58,558,270) |
10526 |
29240 |
20.56 |
49.93 |
|
Egypt (100,388,073) |
26237 |
28615 |
20.12 |
28.5 |
|
Nigeria (200,963,599) |
6407 |
11166 |
7.85 |
5.56 |
|
Algeria (43,053,054) |
1267 |
9733 |
6.84 |
22.61 |
|
Ghana (30,417,856) |
378 |
8548 |
6.01 |
28.1 |
|
Morocco (36,471,769) |
4 |
7778 |
5.47 |
21.33 |
|
Cameroon (25,876,380) |
93 |
5362 |
0.33 |
20.72 |
|
Sudan (42,813,238) |
7 |
4521 |
3.18 |
10.56 |
|
Guinea (12,771,246) |
10 |
3891 |
2.74 |
30.47 |
|
Djibouti (973,560) |
14 |
3779 |
2.66 |
388.16 |
|
Cote d'Ivoire (25,716,544) |
101 |
2477 |
1.74 |
9.63 |
|
Gabon (2,172,579) |
6 |
2238 |
1.57 |
103.01 |
|
Mayotte* (NA) |
NA |
1993 |
1.4 |
NA |
|
DRC (86,790,567) |
58 |
1945 |
1.37 |
2.24 |
|
Somalia (15,442,905) |
3 |
1828 |
1.29 |
11.84 |
|
Mali (19,658,031) |
11 |
1384 |
0.97 |
7.04 |
|
Guinea-Bissau (1,920,922) |
2 |
1178 |
0.83 |
61.32 |
|
Kenya (52,573,973) |
513 |
1097 |
0.77 |
2.09 |
|
Tunisia (11,694,719) |
1 |
1093 |
0.77 |
9.35 |
|
Zambia (17,861,030) |
22 |
1057 |
0.74 |
5.92 |
|
Equatorial Guinea (1,355,986) |
13 |
1043 |
0.73 |
76.92 |
|
South Sudan* (11,062,113) |
NA |
994 |
0.69 |
8.99 |
|
Niger (23,310,715) |
98 |
961 |
0.68 |
4.12 |
|
Madagascar (26,969,307) |
26 |
908 |
0.62 |
3.37 |
|
Senegal (16,296,364) |
121 |
822 |
0.58 |
5.04 |
|
Ethiopia (112,078,730) |
146 |
731 |
0.51 |
0.65 |
|
Chad (15,946,876) |
5 |
700 |
0.49 |
4.39 |
|
CAR (4,745,185) |
6 |
671 |
0.47 |
14.14 |
|
Mauritania (4,525,696) |
3 |
668 |
0.47 |
14.76 |
|
Burkina Faso (20,321,378) |
211 |
662 |
0.47 |
3.26 |
|
Congo* (5,380,508) |
NA |
571 |
0.4 |
10.61 |
|
Tanzania (58,005,463) |
13 |
509 |
0.36 |
0.88 |
|
Cape Verde (549,935) |
4 |
466 |
3.77 |
84.74 |
|
Togo (8,082,366) |
25 |
428 |
0.3 |
5.29 |
|
Uganda (44,269,594) |
18 |
418 |
0.29 |
0.94 |
|
Rwanda (12,626,950) |
54 |
397 |
0.28 |
3.14 |
|
Malawi* (18,628,747) |
NA |
358 |
0.25 |
1.92 |
|
Mauritius (1,265,711) |
93 |
335 |
0.24 |
26.47 |
|
Mozambique (30,366,036) |
7 |
316 |
0.22 |
1.04 |
|
Eswatini (1,148,130) |
9 |
272 |
0.19 |
23.69 |
|
Sierra Leone* (7,813,215) |
NA |
257 |
0.18 |
3.29 |
|
Liberia (4,937,374) |
3 |
200 |
0.14 |
4.05 |
|
Zimbabwe (14,645,468) |
36 |
149 |
0.1 |
1.02 |
|
Comoros* (850,886) |
NA |
132 |
0.09 |
15.51 |
|
Benin (11,801,151) |
5 |
90 |
0.06 |
0.76 |
|
Libya* (6,777,452) |
NA |
73 |
0.05 |
1.08 |
|
Eritrea (3,213,972)Δ |
6 |
39 |
0.03 |
1.21 |
|
Angola (31,825,295) |
4 |
35 |
0.02 |
0.11 |
|
Namibia (2,494,530) |
14 |
26 |
0.02 |
1.04 |
|
Gambia (2,347,706) |
6 |
25 |
0.02 |
1.06 |
|
Botswana* (2,303,697) |
NA |
23 |
0.02 |
0.99 |
|
Burundi (11,530,580) |
NA |
19 |
0.01 |
0.16 |
|
Sao Tome and Principe* (215,056) |
NA |
11 |
0.01 |
5.12 |
|
Seychelles (97,625) |
7 |
11 |
0.01 |
11.27 |
|
Lesotho* (2,125,268) |
NA |
2 |
0.001 |
0.09 |
|
Total |
46593 |
142245 |
||
|
*no data on the COVID-19 Data Working Group line list; †based on WHO-OCHA data; ‡ Data from World Bank database: Health Nutrition and Population Statistics. Last Updated: 07/02/2020; Δ based on 2011 data; CAR=Central African Republic; DRC= Democratic Republic of the Con |
||||
Table 1: Cumulative number of cases of COVID-19 in African Countries up to end of May 2020.
3.2 Number of countries submitting SARS-CoV-2 sequence data
Based on the sequence data from Africa available on GISAID database as of 27 May 2020, we calculated the number of sequences from each country out of the total 227 sequence data available as of that date. Results are shown in Table 2. Only 9 countries from Africa have submitted SARS-CoV-2 genetic sequence data to GISAID as of the date of interest. The bulk of sequence data (n=133; ≈58.6%) were from the Democratic Republic of Congo (DRC) despite not having high number of cases like countries with very high cases at the period. Similarly, the country with second highest number of sequence data, Senegal (n=23; ≈10.1%), had fewer number of cases compared to the top five countries with highest number of cases at this period. This may reflect the molecular sequencing capacity that these countries have developed over the years as part of their involvement in diagnosing other infectious diseases in the continent, especially Ebola in the DRC.
|
Country |
Number of Sequences |
Percent |
|
Democratic Republic of the Congo |
133 |
0.586 |
|
Senegal |
23 |
0.101 |
|
South Africa |
20 |
0.088 |
|
Ghana |
15 |
0.066 |
|
Tunisia |
6 |
0.026 |
|
Algeria |
3 |
0.013 |
|
Gambia |
3 |
0.013 |
|
Egypt |
2 |
0.009 |
|
Nigeria |
2 |
0.009 |
Table 2b: Number of SARS-CoV-2 sequences submitted to GISAID database from Africa as of 27 May 2020.
3.3 Age and gender distribution of cases for which genetic sequence are available
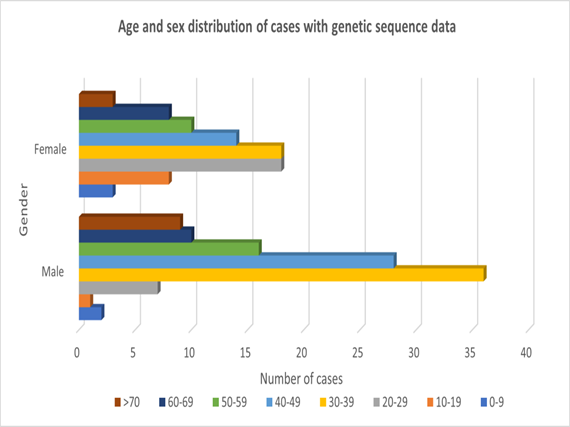
We estimated the median age of cases for which genetic sequences are available and the gender distribution of the cases. Based on the available data, the median age for all represented cases is 38 years (range: 3years to 87 years), of which median age for females is 38.5 years (range: 5 years to 75 years) and that for male is 42.5 years (range: 3 years to 87 years). For cases in which data are available on gender, 82 (42.9%) are female, while 109 (57.1%) are male. The age and gender distribution of the cases is shown in Figure 2. Most of the available sequences are obtained from individuals aged between 30 to 50 years, reflecting the age groups that are more infected with SARS-CoV-2 in Africa. Also, there are more sequence data for men. This supports the well-known observation that men are more affected by COVID-19 than women for obvious reasons.
3.4 Clade distribution of SARS-CoV-2 isolates from Africa
Based on the metadata on GISAID platform, we estimated the number of genetic sequences from Africa belonging to the different GISAID clades classification for SARS-CoV-2. As can be seen from Figure 3, most of the SARS-CoV-2 sequenced in Africa belonged to clade G. The exceptions are Algeria and Egypt in which the available 3 and 2 sequences respectively belonged to clade GH, and Senegal where clade GH slightly outnumber clade G.
3.5 Output of sequence alignment and phylogenetic tree
The original 227 sequences were manually filtered and partial gene sequence with less than 29000bp were excluded. This now resulted in 221 sequences that were subjected to Multiple Sequence Alignments (MSA). Result of MSA produced aligned sequences with Mean sequence length of 29783.9 (S.D=115.6) bp (Min. = 29302bp and Max. = 29903) with base coverage of 29.3%, 18%, 19.3%, and 32.1% for Adenine (A), Cytosine (C), Guanine (G) and Thymidine (T) respectively. About 129,551 (2.0% of overall alignment) positions contained gaps.
Maximum likelihood statistics was applied to the aligned sequence to find the best DNA model with which to build a maximum likelihood tree. Results of 24 model fits were generated (see Table S2) and model was selected based on the lowest BIC score (Bayesian Information Criterion) which is considered to optimally describe the best substitution pattern among the bases. All positions containing gaps and missing data were eliminated.
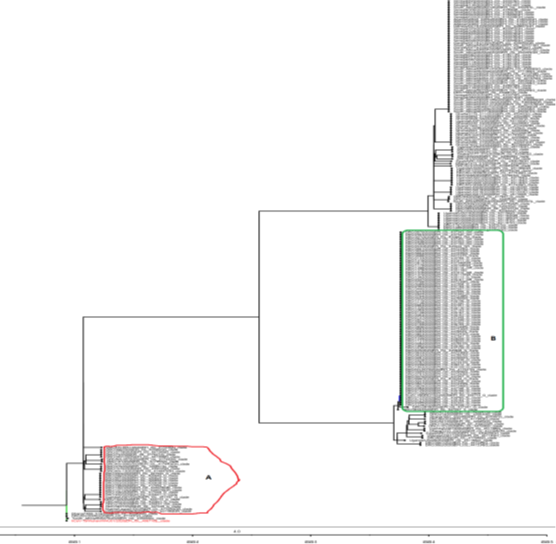
The evolutionary tree obtained (see Figure 4) was inferred using the Maximum Likelihood method and General Time Reversible (GTR) model. Branch length of the tree is based on the number of substitutions per site. The tree reflected the period that Africa began to record cases of COVID-19 and the diversity that the virus has undergone within the continent. The earliest available sequences clustered closely to the reference Wuhan isolate, indicating multiple introductions of virus closely related to the reference strain into the continent from outbreak clusters outside the continent (see area A). However, there is also possibility that the virus was already circulating in the continent even before the first confirmed case was announced on February 15, 2020. This can be inferred from the very close affinity between three virus strains from the continent - Ghana/1659_S14/2020|EPI_ISL_422405, DRC/KN0054/2020|EPI_ISL_417437, and South_Africa/R05475/2020|EPI_ISL_435059 from Ghana, DRC, and South Africa respectively – with the Wuhan reference strain. The clustering of the virus at the longer tips of the tree reflects geographic adaptation of the virus to the new location and local circulation of adapted strains with less evidence of notable viral mutations. Given that most of the isolates are from DRC, there is a nodal cluster of viruses from DRC occupying a tree branch (see area B). This reflects ongoing local transmission within the country as the effect of lockdown imposed in most countries might not have allowed importation of cases from outside the country. Generally, the viral divergence has remained stable within the continent and within individual countries since the earlier multiple introductions from different sources from outside the continent.
4. Discussion
The COVID-19 pandemic, though abated through vaccine introduction, has however remained since the world awoke to the knowledge of a novel strain of coronavirus in the twilight of the 2020 decade. Effort to control it has been met with mixed successes and failures, with countries like China, hailed to have made commendable strides in tackling it at the initial stage latter facing waves of resurgent outbreaks [23].
In this work, we employed data generously made available by scientists from different laboratories around the world to the GISAID initiative to interrogate the genetic epidemiology of the novel virus in Africa. This was intended among others to understand how outbreaks in Africa, which were seeded from other continents, especially Europe [8], have interacted to produce the pattern predominant at that time in Africa. And help to understand viral divergence occurring in a location that could be important for other locales in monitoring imported cases. Although our analysis did not include viruses outside Africa, the result obtained however provided certain insight into how the virus adapted in Africa after multiple introductions from outside the continent at the onset of the outbreak in the continent [14,25].
Our results showed that the number of cases of COVID-19 increased progressively after the first case was detected in Egypt on February 15, 2020. The number of available sequence data submitted to GISAID was however not proportionate to the number of cases in different countries. Most of the sequence data on GISAID platform from Africa were from the DRC. This could be a reflection of the genetic diagnostic capability the country developed following its long battle with the Ebola virus [26]. Also, the median age of 38 years (range: 3years to 87 years) for cases in which there were sequence data was in agreement with the median age of the infected of 36 years reported by the World Health Organisation [4]. Moreover, the preponderance of clade G in the sequences from Africa agreed with the observation of O'Toole (27) with respect to sequences from DRC.
Phylogenetic analysis of the sequence data supported evidence for multiple introduction into Africa of SARS-CoV-2 strains from different continents at the beginning of the pandemic in Africa [8,14,25]. This is similar to observations in other continents too [24,28]. However, the affine clustering of some viruses from Africa - Ghana/1659_S14/2020|EPI_ISL_422405, DRC/KN0054/2020|EPI_ISL_417437, and South_Africa/R05475/2020|EPI_ISL_435059 from Ghana, DRC and South Africa respectively - to the reference Wuhan strain, could suggest an earlier circulation of the virus in the continent prior to the reporting of the first case in February 15 2020. In any case, we could not be highly confident in this conclusion as we do not have access to a comprehensive epidemiological data detailing the travel and medical history of the cases in which these viruses were isolated. Generally, our results showed that the viruses circulating in the continent remained stable both within the continent and within individual countries since the earlier multiple introductions from different sources outside the continent.
There are certain limitations that could affect the conclusions of this study. Firstly, there was underreporting of cases of COVID-19 in Africa both from official and unofficial sources [29]. This was clearly reflected in one of our data sources. Secondly, we did not undertake critical evaluation of nucleotide changes in the viruses employed in the study. Thus, any inference to genetic diversity or viral adaptation had to be made with caution. Thirdly, we did not include viruses from other continents apart from the reference Wuhan strain that was used as ancestral lineage. Therefore, we were constrained to make categorical statement on the exact origin (s) of the virus circulating in Africa. Lastly, there was seeming intra-continental mixing in the circulating strains of the virus. However, we could not make a conclusive statement on this because we do not have detailed information on travel and medical history of persons in which the viruses were isolated.
5. Conclusion and Recommendation
The current study was conceptualized to evaluate the genetic epidemiology of COVID-19 infection in Africa by analysing sequence data submitted to GISAID database from laboratories in Africa. The original intention also included linking information on GISAID database with those contained in the data curated by the COVID-19 Open Data Working Group. The latter could not be done because certain key data points were lacking in the curated data. Also, the apparent underreporting of the number of infections in Africa in the data from COVID-19 Open Data Working Group warranted a comparison with data from another source (WHO-OCHA). While the results of other demographically related data agrees with what has been reported elsewhere [4], the outcome of this study however provided evidence to support multiple introductions of SARS-CoV-2 into Africa, and further suggested that the virus may have already been circulating in the continent prior to official recording of the first case in the continent. Moreover, there was strong impression to infer certain genetic adaptation of the virus in the continent that informed the close clustering of less distant isolates from the continent. Therefore, it is important for public health authorities to keep monitoring the genetic sequence data for early detection of unique mutations or external introduction of ‘foreign strain’ into the continent. Other researchers should undertake in-depth profiling of the genetic sequences to detect any nucleotide changes that may infer geographical adaptation. This will go a long way in developing interventions that are not generic but tailored to the need of the continent.
Acknowledgement
The authors are grateful to the following persons and institution for their assistance:
Xin Chen (Jessie), for tutorial assistance and support in producing phylogenetic tree. Aye Moa of the Biosecurity Research Program in The Kirby Institute for her excellent organisation and coordinating capacity. The Australia Awards Scholarship programme for their scholarship funding to CJN.
References
- Novel Coronavirus (2019-nCoV) Situation Report – 1-21 January 2020. Geneva, Switzerland: WHO, 2020 20/01/2020. Report No.: 1 Contract No: 5 (2020).
- Gorbalenya AE, Baker SC, Baric RS, et al. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology 5 (2020): 536-544.
- Naming the coronavirus disease (COVID-19) and the virus that causes it (2020).
- COVID-19: Situation update for the WHO African Region - 1 July 2020. Brazzavile: World Health Organization, Regional Office for Africa (2020).
- Li Q, Guan X, Wu P, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. New England Journal of Medicine 382 (2020): 1199-1207.
- Coronavirus disease (COVID-19) Situation Report – 181. Geneva, Switzerland: World Health Organization (2020).
- Africa CDC. Outbreak Brief #27: Coronavirus Disease 2019 (COVID-19) Pandemic. Africa Centre for Disease Control and Prevention, 2020 21 July (2020).
- Makoni M. Africa Contributes SARS-CoV-2 Sequencing to COVID-19 Tracking. THE SCIENTIST (2020).
- Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data – from vision to reality. Eurosurveillance 22 (2017).
- Llanes A, Restrepo CM, Caballero Z, et al. Betacoronavirus Genomes: How Genomic Information has been Used to Deal with Past Outbreaks and the COVID-19 Pandemic. International Journal of Molecular Sciences 21 (2020): 4546.
- Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34 (2018): 4121-4123.
- Bedford T, Neher R, Hadfield J, et al. Nextstrain: Real-time tracking of pathogen evolution (2020).
- Situation Report Hiatus (2020).
- Oluniyi P. SARS-CoV-2 Genomes from Nigeria Reveal Community Transmission, Multiple Virus Lineages and Spike Protein Mutation Associated with Higher Transmission and Pathogenicity (2020).
- Xu B, Kraemer MUG, Xu B, et al. Open access epidemiological data from the COVID-19 outbreak. The Lancet Infectious Diseases (2020).
- EpiCoVTM: Pandemic coronavirus causing COVID-19 (2020).
- COVID-19 Africa: infected, recovered and diseased [Internet] (2020).
- Health Nutrition and Population Statistics [Internet] (2020).
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32 (2004): 1792-1797.
- Kumar S, Stecher G, Li M, et al. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution 35 (2018): 1547-1549.
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press (2000).
- Rambaut A. FigTree v1.4.4 2006-2018 (2018).
- Coronavirus disease 2019 (COVID-19) Situation Report – 89. Geneva, Switzerland: World Health Organisation, 2020 18/04/2020. Report No (2020).
- Seemann T, Lane CR, Sherry NL, et al. Tracking the COVID-19 pandemic in Australia using genomics. medRxiv (2020).
- Githinji G. Novel 2019 coronavirus Genome Reports [Internet] (2020).
- Palacios G. Ebolavirus [Internet] (2018).
- O'Toole Á. Novel 2019 coronavirus nCoV-2019 Genomic Epidemiology [Internet] (2020).
- AdebalI O, BIrcan A, ÇIrcI D, et al. Phylogenetic analysis of SARS-CoV-2 genomes in Turkey. Turk J Biol 44 (2020): 146-156.
- Mbow M, Lell B, Jochems SP, et al. COVID-19 in Africa: Dampening the storm? Science 369 (2020): 624.


 Impact Factor: * 3.6
Impact Factor: * 3.6 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 