IPSC-Derived Astrocytes to Model Neuroinflammatory and Metabolic Responses in X-linked Adrenoleukodystrophy
Article Information
Parveen Parasar, Navtej Kaur, Jaspreet Singh*
Department of Neurology, Henry Ford Health, Detroit, MI, USA
*Corresponding author: Jaspreet Singh, Department of Neurology, Henry Ford Health, Detroit, MI, USA.
Received: 17 July 2023; Accepted: 24 July 2023; Published: 04 August 2023
Citation: Parveen Parasar, Navtej Kaur, Jaspreet Singh. iPSC-Derived Astrocytes to Model Neuroinflammatory and Metabolic Responses in X-linked Adrenoleukodystrophy. Journal of Biotechnology and Biomedicine. 6 (2023): 281-293.
View / Download Pdf Share at FacebookAbstract
X-linked adrenoleukodystrophy (X-ALD) is an inherited metabolic disorder caused by pathogenic variants in the ABCD1 gene, leading to accumulation of saturated very long chain fatty acids (VLCFA) in body fluids and tissues including brain and spinal cord. In the absence of a clear genotype-phenotype correlation the molecular mechanisms of the fatal cerebral adrenoleukodystrophy (cALD) and the milder adrenomyeloneuropathy (AMN) phenotypes remain unknown. Given our previous evidence of role of astrocytes in the neuroinflammatory response in X-ALD we investigated the metabolic and molecular profiles of astrocytes derived from induced pluripotent stem cells (iPSC). The iPSCs were in turn generated from skin fibroblasts of healthy controls and patients with AMN or cALD. AMN and cALD astrocytes exhibited lack of ABCD1 and accumulation of VLCFA, a biochemical hallmark of X-ALD disease. Accumulation of VLCFA was significantly higher in cALD astrocytes. Mitochondrial function analysis by Seahorse extracellular flux identified increased oxygen consumption and extracellular acidification rates in cALD astrocytes, yet the ATP levels were decreased. Molecular signaling identified increased phosphorylation of STAT3 in cALD astrocytes, and higher proinflammatory cytokine and Toll like receptor (TLR) expression. CRISPR-Cas9 knock-in of functional ABCD1 gene expression differentially affected the expression of key molecular and metabolic targets in AMN and cALD astrocytes. AMN and cALD iPSC-derived astrocytes and their isogenic controls demonstrate differential aspects of X-ALD metabolic and inflammatory response to ABCD1 mutation and can be further utilized for exploring the contribution of iPSC-derived astrocytes to differential X-ALD disease pathology.
Keywords
X-ALD; Astrocytes; iPSC; Neuroinflammation; ABCD1; Mitochondria; Seahorse extracellular flux; AMN; cALD; Fibroblasts, CRISPR/Cas9
X-ALD articles X-ALD Research articles X-ALD review articles X-ALD PubMed articles X-ALD PubMed Central articles X-ALD 2023 articles X-ALD 2024 articles X-ALD Scopus articles X-ALD impact factor journals X-ALD Scopus journals X-ALD PubMed journals X-ALD medical journals X-ALD free journals X-ALD best journals X-ALD top journals X-ALD free medical journals X-ALD famous journals X-ALD Google Scholar indexed journals Astrocytes articles Astrocytes Research articles Astrocytes review articles Astrocytes PubMed articles Astrocytes PubMed Central articles Astrocytes 2023 articles Astrocytes 2024 articles Astrocytes Scopus articles Astrocytes impact factor journals Astrocytes Scopus journals Astrocytes PubMed journals Astrocytes medical journals Astrocytes free journals Astrocytes best journals Astrocytes top journals Astrocytes free medical journals Astrocytes famous journals Astrocytes Google Scholar indexed journals iPSC articles iPSC Research articles iPSC review articles iPSC PubMed articles iPSC PubMed Central articles iPSC 2023 articles iPSC 2024 articles iPSC Scopus articles iPSC impact factor journals iPSC Scopus journals iPSC PubMed journals iPSC medical journals iPSC free journals iPSC best journals iPSC top journals iPSC free medical journals iPSC famous journals iPSC Google Scholar indexed journals Neuroinflammation articles Neuroinflammation Research articles Neuroinflammation review articles Neuroinflammation PubMed articles Neuroinflammation PubMed Central articles Neuroinflammation 2023 articles Neuroinflammation 2024 articles Neuroinflammation Scopus articles Neuroinflammation impact factor journals Neuroinflammation Scopus journals Neuroinflammation PubMed journals Neuroinflammation medical journals Neuroinflammation free journals Neuroinflammation best journals Neuroinflammation top journals Neuroinflammation free medical journals Neuroinflammation famous journals Neuroinflammation Google Scholar indexed journals ABCD1 articles ABCD1 Research articles ABCD1 review articles ABCD1 PubMed articles ABCD1 PubMed Central articles ABCD1 2023 articles ABCD1 2024 articles ABCD1 Scopus articles ABCD1 impact factor journals ABCD1 Scopus journals ABCD1 PubMed journals ABCD1 medical journals ABCD1 free journals ABCD1 best journals ABCD1 top journals ABCD1 free medical journals ABCD1 famous journals ABCD1 Google Scholar indexed journals Mitochondria articles Mitochondria Research articles Mitochondria review articles Mitochondria PubMed articles Mitochondria PubMed Central articles Mitochondria 2023 articles Mitochondria 2024 articles Mitochondria Scopus articles Mitochondria impact factor journals Mitochondria Scopus journals Mitochondria PubMed journals Mitochondria medical journals Mitochondria free journals Mitochondria best journals Mitochondria top journals Mitochondria free medical journals Mitochondria famous journals Mitochondria Google Scholar indexed journals Seahorse extracellular flux articles Seahorse extracellular flux Research articles Seahorse extracellular flux review articles Seahorse extracellular flux PubMed articles Seahorse extracellular flux PubMed Central articles Seahorse extracellular flux 2023 articles Seahorse extracellular flux 2024 articles Seahorse extracellular flux Scopus articles Seahorse extracellular flux impact factor journals Seahorse extracellular flux Scopus journals Seahorse extracellular flux PubMed journals Seahorse extracellular flux medical journals Seahorse extracellular flux free journals Seahorse extracellular flux best journals Seahorse extracellular flux top journals Seahorse extracellular flux free medical journals Seahorse extracellular flux famous journals Seahorse extracellular flux Google Scholar indexed journals AMN articles AMN Research articles AMN review articles AMN PubMed articles AMN PubMed Central articles AMN 2023 articles AMN 2024 articles AMN Scopus articles AMN impact factor journals AMN Scopus journals AMN PubMed journals AMN medical journals AMN free journals AMN best journals AMN top journals AMN free medical journals AMN famous journals AMN Google Scholar indexed journals cALD articles cALD Research articles cALD review articles cALD PubMed articles cALD PubMed Central articles cALD 2023 articles cALD 2024 articles cALD Scopus articles cALD impact factor journals cALD Scopus journals cALD PubMed journals cALD medical journals cALD free journals cALD best journals cALD top journals cALD free medical journals cALD famous journals cALD Google Scholar indexed journals Fibroblasts, CRISPR/Cas9 articles Fibroblasts, CRISPR/Cas9 Research articles Fibroblasts, CRISPR/Cas9 review articles Fibroblasts, CRISPR/Cas9 PubMed articles Fibroblasts, CRISPR/Cas9 PubMed Central articles Fibroblasts, CRISPR/Cas9 2023 articles Fibroblasts, CRISPR/Cas9 2024 articles Fibroblasts, CRISPR/Cas9 Scopus articles Fibroblasts, CRISPR/Cas9 impact factor journals Fibroblasts, CRISPR/Cas9 Scopus journals Fibroblasts, CRISPR/Cas9 PubMed journals Fibroblasts, CRISPR/Cas9 medical journals Fibroblasts, CRISPR/Cas9 free journals Fibroblasts, CRISPR/Cas9 best journals Fibroblasts, CRISPR/Cas9 top journals Fibroblasts, CRISPR/Cas9 free medical journals Fibroblasts, CRISPR/Cas9 famous journals Fibroblasts, CRISPR/Cas9 Google Scholar indexed journals
Article Details
Introduction
X-linked adrenoleukodystrophy (X-ALD) is a progressive peroxisomal disease due to a defective ABCD1 gene encoding the peroxisomal ABC half-transporter ABCD1 or adrenoleukodystrophy (ALD) protein. ABCD1 mutations impair β-oxidation of fatty acids, leading to accumulation of very long chain fatty acids (VLCFAs), predominantly C26:0 in blood and tissues, which adversely affects the brain, spinal cord, and adrenals [1]. A total of 1000 unique variants of ABCD1 in >3400 cases ABCD1 variants have been reported in X-ALD; however, no correlation of genotype to clinical phenotype exists (https://adrenoleukodystrophy.info/) [2]. X-ALD phenotypes include rapidly progressive inflammatory demyelinating cerebral adrenoleukodystrophy (cALD), milder adult-onset forms, adrenomyeloneuropathy (AMN. Approximately 35% of affected males develop cALD before reaching 12 years of age. Without early intervention, most patients with cALD die within a decade after diagnosis. AMN is a gradually developing phenotype, first affecting males with dysfunctional ABCD1 at age 20-30 years manifesting with gait disturbances and bladder dysfunction. About 30% of patients with AMN develop cerebral inflammation, ultimately progressing to the fatal cALD form in adulthood [1, 2]. Human astrocytes are the major cell population in the central nervous system and play a substantial role in X-ALD pathogenesis, and the peroxisomal ABCD1 protein plays a role in the murine astrocyte inflammatory response [3]. We previously documented that loss of adenosine monophosphate-activated protein kinase α1 (AMPKα1) in patients with cALD was associated with a higher inflammatory profile in patient-derived cells [4-6]; however, the underlying definitive mechanisms of phenotypic variability and differential inflammatory responses between AMN and cALD phenotypes remain unknown. Current mouse models [7-9] poorly represent clinical X-ALD phenotypic heterogeneity, and mice do not develop neurologic phenotypes representative of human cALD phenotypes. This begs a clear need for a better cellular model to study X-ALD disease mechanisms. In the context of limited models and inaccessibility of human astrocytes, using induced pluripotent stem cells (iPSCs) from patients is an emerging paradigm to study etiopathogenetic mechanisms at the cellular level in vitro, screen potential therapeutics, and provide defined conditions for reproducibly performing research [10]. Furthermore, iPSCs retain pluripotency, the ability to differentiate into any cell type, and they carry the genetic background of patients, including disease-causing genetic variants [11]. Here we document that reprogrammed iPSCs from healthy control (CTL), AMN, and cALD patient fibroblasts can be differentiated into astrocytes. We document differential inflammatory and metabolic response in iPSC-derived astrocytes from patients with cALD and AMN. These iPSC-derived astrocytes provide a valuable model to investigate differential disease mechanisms in AMN and cALD. Furthermore, CRISPR/Cas9 knock-in of ABCD1 gene in AMN and cALD astrocytes led to differential reversal of mitochondrial, glycolytic, and inflammatory profile in cALD and AMN astrocytes highlighting the role of pathways beyond ABCD1 deletion.
Materials, methods, and Data Analysis
Ethics Approval
The project was approved by Henry Ford Health IRB (#13352). Fibroblast samples were previously de-identified specimens and did not involve recruitment of human subjects.
Human fibroblasts
Human skin fibroblast samples healthy CTL fibroblasts (GM03377; 19-year-old, male), AMN fibroblasts (GM07530; 26-year-old, male), and cALD (GM04933; 8-year-old, male) were obtained from the National Institute for General Medical Sciences human genetic cell repository at Coriell Institute for Medical Research, Camden, NJ.
Derivation of iPSCs and Differentiation into Astrocytes
iPSCs were generated from skin fibroblasts with the Cytotune2.0 Sendai Virus Reprogramming Kit (Axol Biosciences Ltd). Neural Stem Cells (NSCs) were generated from human induced pluripotent stem cells (hiPSCs) via dual SMAD inhibition[12]. These were then differentiated to astrocytes in defined conditions[13]. Briefly, the NSCs were patterned to a glial fate for 14 days and then expanded into Astrocyte Differentiation Medium for a further 21 days. Finally, immature astrocytes were passaged and expanded into Astrocyte Maturation Medium for further 7 days before cryopreservation. Cells were fed with relevant fresh medium every other day throughout the process.
iPSC-Derived Astrocyte Culture
Cells were cultured at seeding density of 5 x 104 cells/cm2 (day 0) at 37°C and 5% CO2 in a humidified incubator in astrocyte medium (Neurobasal-A medium, Thermo Fisher Scientific) containing N-21 max (1X, R&D Systems), fetal bovine serum One Shot (1X, Thermo Fisher Scientific), Glutamax (1X, Gibco, Thermo Fisher Scientific), heregulin-β1 (10 ng/mL, Peprotech, Inc.), basic fibroblast growth factor (8 ng/mL, R&D Systems), penicillin-streptomycin (1X, Thermo Fisher Scientific). For gene expression analysis, cells were cultured in Neurobasal-A medium without serum on Matrigel-coated culture plates. The fibroblasts were characterized and confirmed by nucleoside phosphorylase, glucose-6-phosphate dehydrogenase, and lactate dehydrogenase isoenzyme electrophoresis at National Institute for General Medical Sciences.
Karyotype Analyses
Human G-banding karyotype analysis was performed using the standard protocol at the Henry Ford Cytogenetics Laboratory. Four metaphases were analyzed per cell type to identify G-banding. Cells were analyzed per the Clinical Cytogenetics Standards and Guidelines[14].
Quantitative Real-Time Polymerase Chain Reaction Gene Expression
Total RNA was extracted with the miRNeasy kit (Qiagen). Single-stranded complementary DNA (cDNA) synthesis and RT-qPCR were conducted using Bio-Rad's CFX96 Real-Time PCR Detection System with Bio-Rad IQ SYBR Green Supermix, as described[3, 5]. Gene expression was normalized to 60S ribosomal L27 gene and samples were run in triplicate (primer sequences are listed in the Supplemental table S1).
VLCFA Analysis
CTL, AMN, and cALD astrocytes (2.5 x 105 cells each) were processed at Wayne State University Lipidomics Core facility. Saturated (hexacosanoic [C26:0], tetracosanoic [C24:0]), monounsaturated hexacosenoic (C26:1), and nervonic acid (C24:1) fatty acids were calculated as ratios against docosanoic (C22:0) fatty acid. Lipids were subject to alkaline methanolysis and resulting fatty acid methyl esters were analyzed by gas chromatography-mass spectrophotometry (QP2010 GC-MS system, Shimadzu Scientific Instruments) equipped with Restek column, as reported[5].
Mitochondrial Oxygen Consumption and Glycolytic Function Measurement
Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured using a Seahorse Bioscience XFe96 Extracellular Flux Analyzer, as described previously[4].
Western Blot Analysis
Cells were homogenized in radioimmunoprecipitation (RIPA) buffer with Halt protease inhibitor cocktail (Thermo Fisher Scientific). 50-100 µg of total protein was electrophoresed as described[5, 6]. Antibodies used are listed in the supplemental table S2.
Transmission Electron Microscopy
One million cells were fixed in 2.5% glutaraldehyde overnight and stained per standard protocol with slight modifications. Cells were kept in Eppendorf tubes before infiltration with 1:1 mixture of propylene oxide and Araldite resin (no rocking). Samples were embedded and cured overnight at 60°C. Embedded samples were sectioned at 120 nm with an EM U67 Ultratome (Leica Microsystems, Inc.) at Department of Pathology, Henry Ford Hospital and collected onto 20 mesh copper grids. Dry grids were stained with uranyl acetate and lead citrate. Images were acquired on a Flash transmission electron microscope equipped with BioSprint camera (Joel USA, Inc.) and image capture software (AMT Imaging Systems).
Reactive Oxygen Species Production
Reactive oxygen species (ROS) were measured with the Abcam assay kit according to manufacturer’s protocol.
Nicotinamide Adenine Dinucleotide Assay
The intracellular nicotinamide adenine dinucleotide/nicotinamide adenine dinucleotide hydrogen (NAD/NADH) assay was performed according to the manufacturer’s protocol (Cayman Chemical).
Mitochondrial DNA content
Genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen). Mitochondrial DNA (mtDNA) content was calculated using quantitative RT-PCR by measuring the threshold cycle ratio (ΔCt) of the mitochondria-encoded gene mtDNA tRNA Leu (UUR) gene and the nuclear β2-microglobulin gene (Table 1)[15]. Data were expressed as mtDNA/nuclear DNA (nDNA).
Enzyme-Linked Immunosorbent Assay
Astrocyte serum-free basal culture supernatants were collected after 72 hours, centrifuged to remove residual cells, and stored at –80°C. Enzyme-linked immunosorbent assay Max Deluxe Set (BioLegend) was used to identify interleukin (IL)-6, monocyte chemoattractant protein-1 (MCP1), IL-1β, tumor necrosis factor (TNF)-α, IL-12p70, and IL-12/IL-23(p40) following manufacturer’s instructions.
Human Phospho-Kinase Proteome Profiler
We used the human proteome profiler array to detect relative phosphorylation of 37 kinases and related total proteins using 250 µg per the manufacturers’ instructions (R&D Systems).
Overexpression of ABCD1 protein
Using CRISPR/Cas9 system, we performed Lipofectamine-mediated (Lipofectamine 2000, Invitrogen) introduction of recombinant ABCD1 transgene. Briefly, we used 5µg of ABCD1 overexpression Lenti-viral plasmid (pLV[Exp]-EGFP:T2A: Puro-EF1A> {hABCD1[NM_000033.4, Vector Builder Inc., Chicago, IL) (Supplemental Fig. S3). 5 µg of plasmid was diluted in 150 µl Opti-MEM and Lipofectamine (10 µl) was diluted in 150 µl Opti-MEM media. After 5 min incubation at RT, the diluted plasmid was mixed with diluted Lipofectamine, and the mixture was incubated for 20 min at RT. 250 µl of the complex was added to 1 million cells cultured in media without antibiotics. The transgene was checked 48-72 hours post-transfection and cells selected with puromycin (0.5ug/mL).
Data Analysis
Data was analyzed using GraphPad Prism software (version 7.0). Normality was assessed with Kolmogorov-Smirnov test. Groups were compared with two-tailed unpaired Student’s t-test for normally distributed or non-parametric Mann-Whitney test for non-normally distributed data. Statistical significance was set at p<0.05.
Results
Generation and Characterization of iPSCs and Differentiated Astrocytes
iPSCs generated from CTL, AMN, and cALD fibroblasts expressed pluripotency markers OCT4, Nanog, Sox2, and TRA-1-60 (Fig. 1A). Astrocytes differentiated from iPSCs expressed multiple mature glial cell markers such as glial fibrillary acidic protein (GFAP), aquaporin 4, TUJ1, S100 calcium-binding protein β (S100-β), aldehyde dehydrogenase family 1 member L1 (ALDH1L1), EAAT1, and were negative for A2B5 (Fig. 1B). ALDH1L1, S100-β, and aquaporin4 are markers of mature astrocytes. Expression of GFAP, S100-β, EAAT1, AQP4, and ALDH1 markers was confirmed by immunoblot and densitometry (Fig. 1C-D). The level of S100-β protein was decreased in cALD and AMN astrocytes compared to CTL. AMN and cALD astrocytes showed higher AQP4 expression (Fig. 1C-D).
Karyotype Analyses
CTL, AMN, and cALD astrocytes displayed normal male karyotypes (Fig. 1E-G) and G-banding patterns (data not shown) with no structural or numerical changes, suggesting that reprogramming and differentiation did not induce chromosomal abnormalities in CTL, AMN and cALD astrocytes.
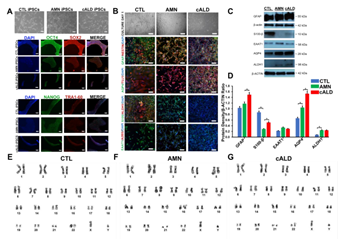
Figure 1: Generation of iPSCs and differentiation into astrocytes. (A) Representative phase contrast and immunostained images of CTL, AMN, and cALD iPSCs. Upper panel shows morphology in culture (day 3) of reprogrammed CTL, AMN, and cALD iPSCs. Middle and lower panel represent phase contrast images for immunostaining for pluripotency markers: OCT4 (green), SOX2 (red), NANOG (green), and TRA1-60 (red) DAPI (blue). (B) Phase contrast images (upper panel) of differentiated CTL, AMN, and cALD astrocytes in culture (day 1) and immunofluorescence staining (lower panels) with astrocyte specific markers: GFAP/Nestin (green/red), S100-β/ALDH1 (green/red), AQP4/tubulin βIII (green/red), and EAAT1/A2B5 (green/red) Nuclei are stained with DAPI. (C) Representative Western blots for GFAP, S100-β, EAAT1, AQP4, ALDH1, and β-actin in CTL, AMN, and cALD astrocytes. (D) Quantification of GFAP, S100-β, EAAT1, AQP4, and ALDH1 (ratio against β-actin) in CTL, AMN, and cALD astrocytes (n=3). (E-G) Karyotype analysis for differentiated astrocytes. AMN: Adrenomyeloneuropathy; cALD: Cerebral adrenoleukodystrophy; CTL: Control; iPSC: Induced pluripotent stem cells. (scale bars 150 µm).
AMN and cALD Astrocytes Show Loss of ABCD1 Protein
There was complete loss of ABCD1 protein in AMN and cALD astrocytes relative to CTL astrocytes. AMN and cALD astrocytes had a marked compensatory increase in ABCD2 protein. ABCD3 was expressed at low levels in CTL, AMN, and cALD astrocytes (Fig. 2A).
AMN and cALD Astrocytes Accumulated Higher VLCFAs
Loss of ABCD1 results in impaired import of VLCFA into peroxisomes resulting in their accumulation. In line with this, AMN astrocytes displayed significantly higher levels of saturated (C24:0, C26:0) and monounsaturated (C24:1, C26:1) (p<0.01 and p<0.05 respectively) VLCFA than CTL astrocytes (Fig. 2B). cALD astrocytes had further significantly increased saturated (p<0.01) and monounsaturated (p<0.001) VLCFA levels than AMN astrocytes (Fig. 2B). Absolute levels of saturated and monounsaturated VLCFA also followed the same trend (Fig. S1). ELOVL1 catalyzes synthesis of saturated and monounsaturated VLCFAs [16]. ELOVL expression was significantly higher (p< 0.01) in cALD astrocytes than AMN and CTL astrocytes, and expression between AMN and CTL astrocytes did not differ. Expression of ELOVL3 did not differ significantly between the three groups (Fig. 2C).
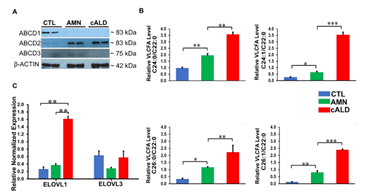
Figure 2: CTL, AMN, and cALD patient astrocytes retain biochemical characteristics of X-ALD. (A) Representative Western blots for ABCD1, ABCD2, and ABCD3, and β-actin in CTL, AMN, and cALD astrocytes. (B) Quantification of VLCFA (C24:0/C22:0 and C24:1/C22:0; C26:0/C22:0 and C26:1/C22:0) in CTL, AMN, and cALD astrocytes. (C) RT-qPCR quantification of ELOVL1 and ELOVL3. The gene expression of the individual sample was assessed with fold change using the comparative ΔΔCt method normalized to L27. Results represent means ± standard error. Two-tailed Student’s t test was used for data analysis. **p<0.01;***p<0.001. VLCFA: Very long chain fatty acids.
Mitochondrial extracellular flux rates in cALD and AMN iPSC-derived astrocytes
VLCFA accumulation is associated with mitochondrial dysfunction in X-ALD [4-6]. The Cell Mito Stress Test, a measure of mitochondrial OCR, showed increased (p<0.001) non-mitochondrial OCR (Fig. 3A-3B), ATP-linked OCR (Fig. 3C), maximal respiration, and spare capacity (Fig. 3D) in cALD astrocytes than AMN astrocytes. ATP rate assay revealed significantly lower ATP production (p<0.05) in AMN and cALD astrocytes than CTL astrocytes, but cALD astrocytes had a significantly higher ATP production rate (p<0.05) than AMN astrocytes (Fig. 3E). We observed elevated glycolytic response in cALD astrocytes, measured as ECAR (Fig. 3F). Glycolysis stress test revealed higher (p<0.001) non-glycolytic ECAR (Fig. 3G) and significantly higher glycolytic capacity (p<0.05) and glycolytic reserve (p<0.05) in cALD astrocytes (Fig. 3H-I) than in AMN astrocytes. While cALD astrocytes had a significantly higher glycolytic ATP (p<0.05) production rate than AMN, both AMN and cALD astrocytes had significantly lower glycolytic ATP production (p<0.05) rates than CTL astrocytes (Fig. 3J).
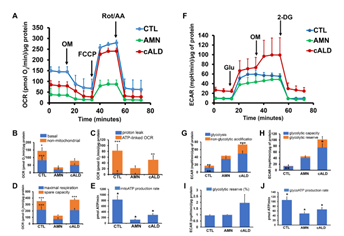
Figure 3: cALD astrocytes show altered parameters of OCR and ECAR in cell mito stress and glycolytic stress test assays. (A) Plot showing overall OCR in CTL, AMN, and cALD astrocytes. (B) Quantification of basal and non-mitochondrial OCR. (C) Quantification of proton leak and ATP-linked OCR. (D) Quantification of maximal respiration and spare respiratory capacity. (E) Quantification of mitochondrial ATP production rate (pmol ATP/minute). (F) Plot showing the overall ECAR in CTL, AMN, and cALD astrocytes. (G) Quantification of glycolysis and non-glycolytic acidification. (H) Quantification of glycolytic capacity and glycolytic reserve. (I) Quantification of percent glycolytic reserve. (J) Quantification of glycolytic ATP production rate (pmol ATP/minute). Results represent means ± standard error. Analysis was performed using the Wave software (version 2.6.1.53; Agilent) and normalized to the cell protein content (A-D and F-I). Data analysis in Figure E and J was performed after acquisition of data using XF Real-Time ATP rate assay generator (Agilent). Unpaired two-tailed Student’s t test or Mann-Whitney test were used for analysis. **p<0.01;***p<0.001. Significance level over AMN bars represent difference between CTL and AMN astrocyte samples and that over cALD bars represent the difference between AMN and cALD astrocyte samples. ECAR: Extracellular acidification rate; OCR: Oxygen consumption rate; OM: Oligomycin; FCCP: Carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone; Rot/AA: Rotenone and antimycin-A; Glu: Glucose; OM: Oligomycin; 2-DG: 2-deoxi-D-glucose.
Electron Microscopy Reveals Mitochondrial Structure Differences
To determine if mitochondrial functional changes were also associated with structural alterations, we performed electron microscopy to assess differences in cellular and mitochondrial structure of CTL, AMN and cALD astrocytes. AMN and cALD astrocytes contained more fatty acid droplets than CTL astrocytes (Fig. 4A1-A3). We also observed disrupted cristae structure in cALD astrocytes relative to CTL and AMN astrocytes (Fig. 4A4-A9). Dynamin related protein-1 (DRP-1), a key mitochondrial fission regulator, was elevated (p<0.01) in both AMN and cALD astrocytes (Fig. 4B-C). These findings suggest that mitochondrial structure perturbations may contribute to mitochondrial dysfunction in X-ALD.
Mitochondrial and Glycolytic Gene expressions are altered in AMN and cALD astrocytes
We performed RT-qPCR to assess if the altered OCR and ECAR was associated with alterations in expression of oxidative phosphorylation (OXPHOS) and glycolytic genes. Succinate dehydrogenase complex subunit B (SDHB) and ubiquinol-cytochrome C reductase complex ubiquinone-binding protein QP-C (UQCRQ) expression were significantly lower in AMN (p<0.01) and cALD (p<0.05) astrocytes (Fig. 4D). cALD astrocytes showed significantly higher (p<0.01) expression of NDUFA6 (NADH:Ubiquinone Oxidoreductase Subunit A6), ATP5B, citrate synthase, nuclear respiratory factor 1, and peroxisome proliferator-activated receptor gamma coactivator 1-α (PGC1α) than AMN astrocytes (Fig. 4D). AMN astrocytes expressed higher citrate synthase (p<0.05) than CTL astrocytes (Fig. 4D). Immunoblot analysis revealed decreased SDHB (Complex II-SDHB) and mitochondrially encoded cytochrome C oxidase I (Complex IV-MTCO1) in cALD astrocytes than in AMN astrocytes, whereas AMN astrocytes had higher levels of SDHB and MTCO1 than CTL astrocytes (Fig. 4E). In line with higher ECAR levels, cALD astrocytes displayed significantly higher expression of glycolytic genes Glut-1/SLC2A, HK-I, HK-II, GPI, ALDOC, GAPDH, ENO-1, and PKM2 than AMN and CTL astrocytes, whereas LDHA was not significantly different among the three groups. PKM2 (p<0.01) and GLUT1 (p<0.01) which were significantly increased in AMN astrocytes relative to CTL astrocytes (Fig. 4F-G).
NADPH oxidase expression is increased in cALD astrocytes
Oxidative damage of proteins is observed in postmortem cALD brain and VLCFA accumulation is associated with oxidative stress. We measured ROS levels in CTL, AMN and cALD astrocytes. ROS generation did not significantly differ between the CTL, AMN and cALD under basal conditions (Fig. 4H) despite increased NADPH oxidase (NOX) isoforms NOX1 (p<0.01), NOX2 (p<0.05) and NOX3 (p<0.01) expression in cALD astrocytes (Fig. 4I). Tert-butyl hydroperoxide (TBHP) was used to induce ROS in positive wells.
AMN and cALD Astrocytes Show Reduced mtDNA Content
To determine if functional mitochondrial changes were associated with changes in mitochondrial content, we determined the mtDNA/nDNA ratios by RT-qPCR. mtDNA/nDNA ratio in AMN astrocytes was significantly lower (p<0.05) than CTL astrocyte (Fig. 4J). cALD astrocytes had further significantly lower (p<0.05) mtDNA/nDNA ratio compared to AMN astrocytes suggesting lower mitochondrial content in X-ALD cells (Fig. 4J).
Intracellular NAD/NADH Levels are Increased in AMN and cALD Astrocytes
NAD is a cofactor involved in electron transfer and NAD/NADH ratio reflects the intracellular redox state. cALD astrocytes showed a significantly higher NAD/NADH ratio (p<0.001) than AMN astrocytes, which in turn had a significantly higher NAD/NADH ratio (p<0.001) than CTL astrocytes (Fig.4K).
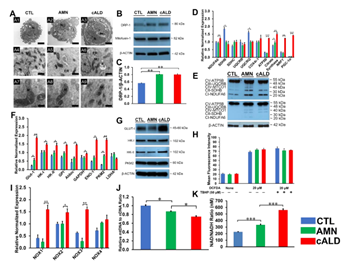
Figure 4: Mitochondrial structural and functional assessment using TEM and RT-qPCR.
(A) Representative TEM images of CTL, AMN, and cALD astrocytes. Upper panel (A1-A3 at scale bars 2 µm) represent whole CTL, AMN, and cALD astrocyte. Middle (A4-A6) and lower (A7-A9) represent mitochondrial structures in CTL, AMN, and cALD astrocytes (scale bars 1 µm and 400 nm, respectively). N: Nucleus; M: Mitochondria; ER: Endoplasmic reticulum; L: Lipid droplets. (B) Western blotting with DRP-1, mitofusin-1, and β-actin in CTL, AMN, and cALD astrocytes. (C) Quantification of DRP-1 (ratio against β-actin) (n=3). (D) RT-qPCR quantification of gene expression for mitochondrial genes (NDUFA6, SDHB, SDHC, UQCRB, UQCRQ, Cox4i-1, ATP5B, citrate synthase, and PGC1α). (E) Representative Western blots for total OXPHOS at low exposure (bottom blot) and high exposure (top blot) and β-actin. (F) RT-qPCR quantification of glycolytic genes (Glut-1, HK-I, HK-II, GPI, ALDOC, GapDH, EN-1, PKM2, and LDHA). (G) Representative Western blots for Glut-I, HK-I, HK-II, PKM2, and β-actin in CTL, AMN, and cALD astrocytes. (H) Quantification of reactive oxygen species (mean fluorescence intensity) in CTL, AMN, and cALD. Tert-butyl hydroperoxide (TBHP) was used to induce ROS in positive wells. (I) RT-qPCR quantification of NOX1, NOX2, NOX3, and NOX4 for CTL, AMN, and cALD astrocytes. (J) mtDNA to nDNA ratio and (K) NAD/NADH ratio in CTL, AMN, and cALD astrocytes. The values represent mean means ± standard error. The gene expression of the individual sample was assessed with fold change using the comparative ΔΔCt method and normalized to L-27. Unpaired two-tailed Student’s t test or Mann-Whitney test were used for analysis. **p<0.01; ***p<0.001. DRP-1: Dynamin related protein-1; mtDNA: Mitochondrial DNA; nDNA: Nuclear DNA; TEM: Transmission electron microscopy.
Proinflammatory, Anti-inflammatory, Chemokine and neurotrophic Gene Expressions
We and others previously documented increased proinflammatory gene expression in postmortem brain tissue from patients with cALD [5]. VLCFA accumulation is associated with the inflammatory response in ABCD1-silenced mouse astrocytes [3]. Proinflammatory gene expression analysis in unstimulated AMN and cALD astrocytes revealed higher interferon-γ (p<0.01), nitric oxide synthase 2, TNFα (both p<0.05), IL-17A (p<0.01), and IL-22 (p<0.01) expression in cALD than AMN astrocytes (Fig. 5A). TNFα expression in AMN was significantly higher (p<0.05) than CTL astrocytes (Fig. 5A). IL-12α (p<0.01) and IL-12β (p<0.05) expression was lower in cALD astrocytes compared with AMN and CTL astrocytes (Fig. 5A). IL-12α expression in AMN was comparable to CTL astrocytes while IL-12β was significantly decreased (p<0.01) (Fig. 5A). AMN astrocytes had higher (p<0.01) IL-6 expression than cALD and CTL astrocytes (Fig. 5A). IL-1β (p<0.05) and IL-RA (p<0.01) expression were decreased in both AMN and cALD astrocytes compared with CTL (Fig. 5A), although IL-1R was significantly (p<0.05) higher in AMN and cALD astrocytes than in CTL astrocytes. IL-1β and IL-23α expressions were comparable between AMN and cALD astrocytes (Fig. 5A). Anti-inflammatory cytokines arginase-1 (ARG1) and mannose receptor C-type1 (MRC1) expression was significantly lower (p<0.05) in cALD astrocytes than in AMN astrocytes (Fig. 5B). ARG1 in AMN astrocytes was in turn significantly lower (p<0.05) than CTL astrocytes. CTL astrocytes had significantly higher (p<0.05) IL-4 levels compared to both AMN and cALD astrocytes. IL-4 levels were comparable between AMN and cALD astrocytes. Fizz1 expression was higher (p<0.05) in cALD astrocytes relative to AMN astrocytes. IL-10 expression was significantly lower (p<0.05) in AMN astrocytes than CTL astrocytes and significantly higher (p<0.01) in cALD astrocytes than in AMN astrocytes. Oncostatin M (OSM) was significantly higher (p<0.05) in AMN compared to CTL and cALD astrocytes (Fig. 5B). AMN astrocytes demonstrated significantly lower (p<0.05) expression of BDNF compared with CTL astrocytes, which was further significantly decreased (p<0.05) in cALD astrocytes (Fig. 5C). CNTF expression was significantly higher in cALD (p<0.01) than in AMN and CTL astrocytes, and no significant change in GDNF levels were seen between the three groups. Both AMN and cALD astrocytes had significantly lower expression of NGF (p<0.05) and significantly higher (p<0.05) expression of NTF3 and NTF4/5 compared with CTL astrocytes (Fig. 5C).
Enzyme-Linked Immunosorbent and Nitrite Assay
AMN astrocyte supernatant contained significantly higher (p<0.05) levels of IL-12p40 than CTL and cALD astrocytes (Fig. 5D). Both IL-6 and MCP1 were significantly higher (p<0.01) in AMN astrocytes than in CTL and cALD astrocytes (Fig. 5D). IL-6 in cALD astrocytes was significantly lower (p<0.05) than in AMN astrocytes, whereas TNFα and IL-1β were undetectable in supernatants of all astrocytes (data not shown).
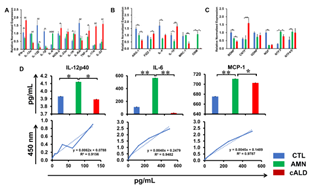
Figure 5: cALD astrocytes reveal an increased proinflammatory cytokine expression RT-qPCR quantification for (A) proinflammatory genes, (B) anti-inflammatory genes and (C) neurotrophic factor genes in CTL, AMN, and cALD astrocytes. The gene expression of the individual sample was assessed with fold change using the comparative ΔΔCt normalized to L-27. (D) Quantification of IL-12p40, IL-6, and MCP-1 in the culture supernatant of CTL, AMN, and cALD astrocytes. Results represent means ± standard error. Two-tailed Student’s t test or Mann-Whitney tests were performed to analyze the data. *p<0.05,**p<0.01;***p<0.001. OD: Optical density (absorbance).
cALD Astrocytes have Increased STAT3 (Serine727) Phosphorylation and Elevated Toll-Like Receptor (TLR) Expression
To determine the signaling underlying the differential cytokine response in AMN and cALD astrocytes, we utilized a human phospho-kinase array proteome profiler. Stat3 phosphorylation at Serine727 was significantly higher in cALD astrocytes (Fig. 6A), which was confirmed by densitometry (p<0.001; Fig. 6B) and immunoblot analysis (Fig. 6C). We identified increased phosphorylation of Akt, p38α, Ras-dependent extracellular signal-regulated kinase (ERK) 1/2, and stress-activated protein kinases/Jun amino-terminal kinases, as well as reduced phosphorylation of AMPKα1 in cALD astrocytes (Fig. 6D-E). We documented higher expression (p<0.05) of TLR1, TLR3, and TLR5 in AMN astrocytes than CTL astrocytes (Fig. 6F). TLR4 was significantly lower (p<0.05) in AMN than in CTL astrocytes (Fig. 6G). cALD astrocytes had significantly higher expression of TLR1 (p <0.001), TLR2 (p<0.05), TLR5 (p<0.01), TLR6 (p<0.05), TLR7 (p<0.05), TLR8 (p<0.05), TLR9 (p<0.05), and TLR10 (p<0.01) than AMN astrocytes. We observed significantly elevated (p<0.05) expression of TLR adaptor protein myeloid differentiation factor 88 (MyD88) in cALD astrocytes (Fig. 6G) along with significantly higher expression (p<0.05) of NF-κB subunits p52 (NF-κB2) and p65 (RelA) in cALD astrocytes than in AMN astrocytes (Fig. 6G).
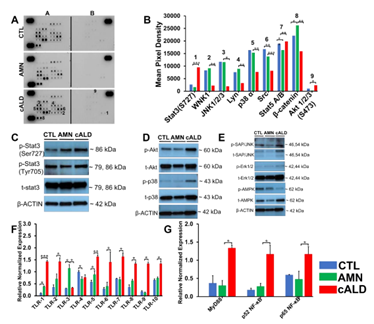
Figure 6: cALD astrocytes show higher Stat3 and TLR expression. (A) Phosphokinase antibody array blots using CTL, AMN, and cALD astrocyte lysates (250 µg for each of antibody array membranes) and (B) Kinase phosphorylation analysis using mean pixel density of Stat3, JNK1/2/3, p38α, SRC, Stat5A/B, β-catenin, and Akt1/2/3 activity in CTL, AMN, and cALD astrocytes. (C) Representative Western blots for pStat3(ser727), pStat3(tyr705), total Stat3, and β-actin. (D) Representative Western blots for phosphorylated and total Akt, p38 and (E) SAP/JNK, Erk1/2, and AMPK in CTL, AMN, and cALD astrocytes. RT-qPCR quantification of TLR1-10 (F) and MyD88, p52 NF-κB, and p65 NF-κB (G). The gene expression of the individual sample was assessed with fold change using the comparative ΔΔCt method normalized to L-27. Results represent means ± standard error. Two-tailed Student’s t test or Mann-Whitney tests were performed to analyze the data. **p<0.01;***p<0.001.
CRISPR-Cas9 knock-in of ABCD1 in AMN and cALD Astrocytes
We next investigated if knock-in of a functional ABCD1 in AMN and cALD astrocytes may reverse the aberrant molecular signature identified above. CRISPR/Cas9 correction of AMN and cALD astrocytes successfully expressed ABCD1 protein in AMN (AMN-OV) and cALD (cALD-OV) astrocytes (Fig. 7A). ABCD1 re-expression significantly reduced the VLCFA (C26:0) levels in AMN and cALD astrocytes (Fig. 7B). Mitochondrial subunit I (NDUFA6) and subunit II (SDHB) were upregulated in AMN-OV and cALD-OV (Fig. 7C). Stat3 phosphorylation at Serine727 and Tyrosine705 was further increased in AMN-OV and cALD-OV indicating that ABCD1 induction did not reverse the Stat3 activity (Fig. 7D). ABCD1 correction significantly increased SDHB (p<0.05), SDHC (p<0.05), UQCRQ (p<0.05), ATP5B (p<0.01), NRF1 (p<0.01), and PGC-1α (p<0.01) in AMN-OV. However, ABCD1 correction reduced (p<0.05) expression of NRF1 and PGC-1α in cALD-OV astrocytes (Fig. 7E). There was no change in NDUFA6, UQCRB, Cox4i1, and Citrate Synthase in AMN-OV and cALD-OV astrocytes (Fig. S2). Glycolytic genes Aldoc and ENO-1 expression was decreased (p<0.05) in AMN-OV astrocytes (Fig. 7F). Expressions of Glut-1, HK-I, HK-II, GPI, Aldoc, ENO-1, PKM2, and LDHA were reduced in cALD-OV astrocytes (ENO-1, p<0.01; all others p<0.05; Fig. 7F). However, Glut-1 and GAPDH were upregulated (p<0.05) in AMN-OV astrocytes (Fig. 7F and Fig. S2). ABCD1 correction significantly mitigated (p<0.01) NOX1 and NOX2 expression in cALD-OV (Fig. 7G) while NOX3 and NOX4 expression was increased (p<0.05; Fig. S2). Interestingly, ABCD1 knock-in significantly lowered the expression of selective proinflammatory genes, IFN-, IL-12α, NOS2, and IL-23α in both AMN-OV and cALD-OV astrocytes whereas IL-1β expression was lowered in AMN-OV only (all p<0.05; Fig. 7H). ABCD1 knock-in resulted in increased (p<0.05) expression of anti-inflammatory genes, Fizz1 and OSM in both AMN-OV and cALD-OV astrocytes (Fig. 7I) but failed to reverse ARG1 expression (Fig. S2). IL-4 (p<0.05), IL-10, (p<0.01) and MRC1 (p<0.05) were upregulated in AMN-OV astrocytes. IL-4 was decreased (p<0.01) and IL-10 and MRC1 remained unchanged in cALD-OV astrocytes (Fig. 7I). There was no change in IL-12β, IL6, IL-1R, IL-RA, and TNFα expression in ABCD1-corrected AMN and cALD astrocytes (Fig. S1). IL-17A (p<0.01) and IL-22 (p<0.05) were significantly increased in AMN-OV astrocytes, however, IL-17A was significantly (p<0.05) reduced in cALD-OV and IL-22 did not change in cALD-OV astrocytes (Fig. S2). ABCD1 knock-in significantly increased expression of BDNF (p<0.01), CNTF (p<0.05), and NGF (p<0.05) in AMN-OV astrocytes whereas only NGF was upregulated in cALD-OV astrocytes (p<0.01). CNTF was significantly downregulated (p<0.05) and BDNF was unchanged in cALD-OV astrocytes (Fig. 7J). No significant change was observed in expressions of GDNF, NTF3, and NTF4/5 in AMN-OV and cALD-OV (Fig. S2). TLR1-2, and TLR5-10 expression were significantly reduced in cALD-OV astrocytes (TLR5 and TLR7 p<0.01; TLR1,2, 6, 8-10, p<0.05); but increased in AMN-OV astrocytes. TLR3 and TLR4 remained unchanged (Fig. 7K and Fig. S2).
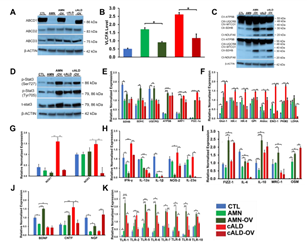
Figure 7: CISPR/Cas9 ABCD1 correction in AMN and cALD Astrocytes. (A) Representative Western blots for ABCD1 and β-actin. (B) RT-qPCR quantification of VLCFA in CTL, AMN, and cALD astrocytes upon ABCD1 induction. (C) Representative Western blots for total OXPHOS at low (bottom blot) and high (top blot) exposure and β-actin. (D) Representative Western blots for pStat3 (ser727), pStat3 (tyr705), total Stat3, and β-actin. RT-qPCR quantification of (E) mitochondrial, (F) glycolytic genes and (G) NADPH Oxidases in CTL, AMN, AMN-OV, cALD, and cALD-OV astrocytes. RT-qPCR quantification of (H) proinflammatory (I) anti-inflammatory and (J) neurotrophic factor genes. (K) RT-qPCR quantification of TLR1, -2, & 5-10. The gene expression of the individual sample was assessed with fold change using the comparative ΔΔCt method and normalized to L-27. Results represent means ± standard error. Two-tailed Student’s t test or Mann-Whitney tests were performed to analyze the data. *p<0.05,**p<0.01;***p<0.001.
Discussion
To date, the mechanistic underpinnings of AMN and cALD phenotypes in X-ALD remain poorly understood. While others have used X-ALD human fibroblast-derived iPSCs to generate brain glial cells [17, 18], in this paper we report key metabolic and molecular differences in the phenotypic profiles of cALD and AMN iPSC-derived astrocytes. Consistent with previous reports, ABCD1 variants in patient fibroblasts did not impede reprogramming and differentiation into astrocytes [17]. iPSC-derived astrocytes from AMN and cALD phenotype not only demonstrated characteristic X-ALD features, including ABCD1 loss and higher VLCFA levels, but also displayed differential mitochondrial, cytokine and signal transduction behavior. VLCFA levels in AMN and cALD patient-derived fibroblasts are not significantly different [3]. However, in line with recent reports, VLCFA accumulation was observed in both AMN and cALD astrocytes with a significantly higher accumulation in cALD astrocytes [3, 19]. Accumulation of VLCFA associated with oxidative stress, affects OXPHOS [20], and induces mitochondrial dysfunction [4-6]. An unexpected finding was increased OCR and ECAR in cALD astrocytes, different from inflammatory situations where normally glycolysis is increased, and OCR is decreased [21]. We previously reported decreased OCR and ECAR in cALD patient-derived fibroblasts [4]. While we have not explored the fate of glutamine, the increased OCR in cALD astrocytes may be in part due to increased availability of glucose-derived pyruvate from higher glycolysis in cALD astrocytes [22]. Seahorse flux analysis also indicated a higher non-mitochondrial OCR, which is associated with enzymes associated with oxidative stress including NADPH oxidases and a higher maximal OCR which may be attributed to higher electron transport capacity in cALD astrocytes. Increased DRP-1 expression in cALD than in AMN astrocytes, suggesting higher mitochondrial fission, and a significant decrease in the mtDNA/nDNA ratio in AMN and cALD astrocytes suggests an overall decreased mitochondrial content. Accumulation of dysfunctional mitochondria can lead to ROS production. Although, expression of superoxide catalyzing NADPH oxidases was increased, we did not detect increased ROS in cALD astrocytes, which may be due to increased antioxidant activities previously reported in X-ALD patient fibroblasts [23]. Increased NAD/NADH ratio is associated with disrupted energy homeostasis, and we observed significantly higher intracellular NAD/NADH ratios in both AMN and cALD astrocytes. A previous study reported a reduced NAD/NADH ratio in ABCD1-silenced human U87 cells [19]. This discordance may be due to the nature of the source of the U87 cells, which are derived from a human glioma patient, compared to AMN and cALD patient-iPSC-derived astrocytes employed in the present study. Moreover, higher NAD/NADH ratios observed in AMN and cALD astrocytes could be due to higher glycolysis, where NADH is oxidized to NAD+. OSM, a late phase cytokine structurally and functionally related to IL-6, is reported to attenuate inflammatory response and limit tissue damage. We observed a synergy is in OSM and IL-6 induction and IL-6 may participate in sustained effects of OSM. Differential IL-6 levels in AMN astrocytes observed between our and a recent study [18] may be due to difference in time of measurement (72h vs 6h in the previous study) or differences in protocols used to generate and differentiate iPSC-derived astrocyte. Moreover, our expression results are also supported by increased IL-6 protein in the supernatant of AMN astrocytes detected by ELISA. IL-6 is also increased in the plasma of AMN patients [24]. OSM induces anti-inflammatory gene expression, ARG1, indirectly through activation of Th2 cytokines in murine macrophages [25]. We observed elevated ARG1 and MRC1 in AMN astrocytes compared to cALD astrocytes which may restrict inflammatory response.
Activated STAT3 may lead to altered mitochondrial energetics [26]. STAT3, can be activated by either the Janus family of kinases or intrinsic tyrosine kinase activity from growth factors and cytokines phosphorylating both serine727 and tyrosine705[27]. Increased phosphorylation of STAT3-p-serine727 and STAT3-p-tyrosine705 indicates that cALD astrocytes may be highly reactive. We previously documented loss of AMPKα1 in cALD fibroblasts, glial cells, and cALD postmortem brain white matter [4-6]. AMPKα1 is the master regulator of energy homeostasis, and its downregulation is associated with neuroinflammatory diseases [28]. Consequently, a higher pSTAT3 and loss of AMPKα1 in cALD astrocytes may underlie the upregulation of cytokine response in cALD astrocytes. mtDNA released by damaged mitochondria due to activated STAT3 [26] may induce endogenous TLR signaling. In line with this, we observed higher expression of TLR1-2, TLR5-10, MyD88, and NF-κB expression in cALD astrocytes. TLR-MyD88-NF-κB pathway is upregulated in the spinal cord of ABCD1-KO mice [29] along with increased NF-κB levels in ABCD1-KO mouse astrocytes [3]. Hematopoietic stem cell transplantation and recently approved gene therapy can arrest cALD disease progression if performed at an early stage [30]. However, the mechanism of disease arrest also remains to be explored since the recovery of lost function remains unfulfilled. We utilized CRISPR/Cas9 knock-in of a functional copy of ABCD1 in AMN (AMN-OV) and cALD (cALD-OV) astrocytes to investigate which of the molecular changes could be rescued by ABCD1. The re-expressed ABCD1 was functional as demonstrated by the reduced VLCFA levels in AMN and cALD astrocytes. ABCD1 induction was sufficient to increase mitochondrial subunits NDUFA6 and SDHB protein levels as seen in OXPHOS immunoblot and expression of SDHB, SDHC, and UQCRQ in AMN astrocytes suggesting that ABCD1 expression can partially restore mitochondrial functions. However, cALD phenotype astrocytes did not show restoration in these gene expressions and thus may require additional therapeutic strategies. ABCD1 knock-in decreased expression of several glycolytic genes in cALD however, expression of key enzymes such as Glut1, GPI, Aldoc and PKM2 were still significantly higher in cALD-OV than CTL or AMN. Similarly, there was selective effect on proinflammatory genes with no change in IL1β, TNFα and IL22 while the expression of IL17A in cALD-OV was still significantly higher than CTL or AMN. More importantly ABCD1 correction could not reverse the expression of anti-inflammatory genes ARG1, IL4 and MRC1 in cALD-OV. ABCD1expression in cALD astrocytes could mitigate the TLR5, TLR7, TLR8 and TLR10 expression. Expression of the other TLRs were either unchanged or were still significantly higher than CTL or AMN. BDNF and CNTF play essential roles in neuronal genesis, differentiation, survival and functional maintenance of sympathetic and parasympathetic neurons [31, 32]. While AMN-OV astrocytes do not show lowering in glycolysis, BDNF, CNTF, and NGF expressions were increased suggesting that ABCD1 induction can enhance neuroprotection in AMN phenotype. On the other hand, ABCD1 induction did not increase BDNF, CNTF, and NGF in cALD astrocytes.
Conclusions
In summary our study provides a point of convergence between mitochondrial aberrations, signal transduction and inflammatory response in AMN and cALD iPSC-derived astrocytes. Human patient iPSC-derived cells could serve to fill the critical gap in understanding of the X-ALD inflammatory pathology and serve as a unique system to identify novel modifiers of inflammatory response in human AMN and cALD. We are aware that the study is based on an in vitro system with limited iPSC-derived lines per phenotype. We also demonstrate for the first time, effect of ABCD1 correction in the corresponding isogenic AMN and cALD astrocytes on mitochondrial function and inflammatory gene expressions. More than 1000 unique variants have been reported in ABCD1 gene with no genotype-phenotype correlation (https://adrenoleukodystrophy.info) and Abcd1-KO mice do not develop the human cALD phenotype. Comparison within the isogenic pair may reduce the phenotypic variation across different iPSC lines from individuals with diverse genetic backgrounds. Our laboratory is working towards further expanding the repertoire of iPSC-derived lines from different X-ALD phenotypes and differentiating them into astrocytes and other neuronal cell types for further investigation of complex interactions.
Data and code availability
The data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Funding
The study was supported by National Institute of Health grants (R01 NS114775 and R21 NS114245) to JS and Funds from Henry Ford Hospital (A10263) to JS.
Authorship contribution statement
JS conceived the idea, designed the experiments, provided direction and funding. PP and NK designed and performed the experiments, contributed to the acquisition and analyses of data. PP drafted the manuscript with contributions from NK and JS. All authors revised and approved the final version of the manuscript.
Acknowledgments
The authors thank Amy Kemper for Electron Microscopy, and Dr. Brandon Shaw and Samantha Turpen for karyotyping. We thank Stephanie Stebens, for formatting the manuscript.
Declaration of competing interest
The authors declare that they have no competing interests to disclose.
References
- HW Moser. Adrenoleukodystrophy. Curr Opin Neurol 8 (1995): 221-226.
- S Kemp, A Pujol, HR Waterham, et al. ABCD1 mutations and the X-linked adrenoleukodystrophy mutation database: role in diagnosis and clinical correlations. Human mutation 18 (2001): 499-515.
- J Singh, M Khan, I Singh. Silencing of Abcd1 and Abcd2 genes sensitizes astrocytes for inflammation: Implication for X-adrenoleukodystrophy. Journal of Lipid Research 50 (2009): 135-147.
- J Singh, S Giri. Loss of AMP-activated protein kinase in X-linked adrenoleukodystrophy patient-derived fibroblasts and lymphocytes. Biochemical and Biophysical Research Communications 445 (2014): 126-131.
- J Singh, B Olle, H Suhail, et al. Metformin-induced mitochondrial function and ABCD2 up-regulation in X-linked adrenoleukodystrophy involves AMP-activated protein kinase. Journal of Neurochemistry 38 (2016): 86-100.
- J Singh, H Suhail, S Giri. Loss of AMP-activated protein kinase induces mitochondrial dysfunction and proinflammatory response in unstimulated Abcd1-knockout mice mixed glial cells. Mediators of Inflammation 2015 (2015): e176983.
- JF Lu, AM Lawler, PA Watkins, et al. A mouse model for X-linked adrenoleukodystrophy. Proceedings of the National Academy of Sciences of the United States of America 94 (1997): 9366-9371.
- S Forss-Petter, H Werner, J Berger, et al. Targeted inactivation of the X-linked adrenoleukodystrophy gene in mice. Journal of neuroscience research 50 (1997): 829-843.
- T Kobayashi, N Shinnoh, A Kondo, et al. Adrenoleukodystrophy protein-deficient mice represent abnormality of very long chain fatty acid metabolism. Biochemical and biophysical research communications 232 (1997): 631-636.
- MC Marchetto, KJ Brennand, LF Boyer, et al. Induced pluripotent stem cells (iPSCs) and neurological disease modeling: progress and promises. Human molecular genetics 20 (2011): R109-R115.
- IH Park, R Zhao, JA West, et al. Daley, Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451 (2008): 141-146.
- Y Shi, P Kirwan, J Smith, et al. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci 15 (2012): 477-486, S471.
- DL Lundin A, Clausen M, Ricchiuto P, et al. Human iPS-Derived Astroglia from a Stable Neural Precursor State Show Improved Functionality Compared with Conventional Astrocytic Models. Stem Cell Reports 13 (2018): 1030-1045.
- FM Mikhail, NA Heerema, KW Rao, et al. Section E6.1-6.4 of the ACMG technical standards and guidelines: chromosome studies of neoplastic blood and bone marrow-acquired chromosomal abnormalities. Genet Med 18 (2016): 635-642.
- V Venegas, J Wang, D Dimmock, et al. Real-time quantitative PCR analysis of mitochondrial DNA content. Curr Protoc Hum Genet 68 (2011): 11-12.
- Y Ohno, S Suto, M Yamanaka, et al. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci USA 107 (2010): 18439-18444.
- J Jang, HC Kang, HS Kim, et al. Induced pluripotent stem cell models from X-linked adrenoleukodystrophy patients. Ann Neurol 70 (2011): 402-409.
- M Baarine, M Khan, A Singh, et al. Functional Characterization of IPSC-Derived Brain Cells as a Model for X-Linked Adrenoleukodystrophy. 10 (2015): e0143238-e0143238.
- BC Baarine M, Singh A, Singh I. ABCD1 deletion-induced mitochondrial dysfunction is corrected by SAHA: implication for adrenoleukodystrophy, J Neurochem 133 (2015): 380-396.
- S Fourcade, J Lopez-Erauskin, J Galino, et al. Pujol, Early oxidative damage underlying neurodegeneration in X-adrenoleukodystrophy. Hum Mol Genet 17 (2008): 1762-1773.
- W Xiao, WM Oldham, C Priolo, et al. Immunometabolic Endothelial Phenotypes: Integrating Inflammation and Glucose Metabolism. Circ. Res 129 (2021): 9-29.
- SC Pamies D, Schvartz D, González-Ruiz V, et al. Neuroinflammatory Response to TNFα and IL1β Cytokines Is Accompanied by an Increase in Glycolysis in Human Astrocytes In Vitro. Int J Mol Sci 22 (2021): 4065.
- CR Vargas, M Wajner, LR Sirtori, et al. Evidence that oxidative stress is increased in patients with X-linked adrenoleukodystrophy. Biochim Biophys Acta 1688 (2004): 26-32.
- M Ruiz, M Jove, A Schluter, et al. Altered glycolipid and glycerophospholipid signaling drive inflammatory cascades in adrenomyeloneuropathy. Hum Mol Genet 24 (2015): 6861-6876.
- A Dubey, L Izakelian, EA Ayaub, et al. Separate roles of IL-6 and oncostatin M in mouse macrophage polarization in vitro and in vivo. Immunol Cell Biol 96 (2018): 257-272.
- J Wegrzyn, R Potla, YJ Chwae, et al. Function of mitochondrial Stat3 in cellular respiration. Science (New York, N.Y.), 323 (2009): 793-797.
- Z Wen, Z Zhong, JE Darnell Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82 (1995): 241-250.
- N Nath, M Khan, R Rattan, et al. Loss of AMPK exacerbates experimental autoimmune encephalomyelitis disease severity. Biochem Biophys Res Commun 386 (2009): 16-20.
- A Schlüter, J Sandoval, S Fourcade, et al. Epigenomic signature of adrenoleukodystrophy predicts compromised oligodendrocyte differentiation. Brain Pathology 28 (2018): 902-919.
- E Shapiro, W Krivit, L Lockman, et al. Long-term effect of bone-marrow transplantation for childhood-onset cerebral X-linked adrenoleukodystrophy. Lancet 356 (2000): 713-718.
- Y Li, F Li, D Qin, et al. The role of brain derived neurotrophic factor in central nervous system. Front Aging Neurosci 14 (2022): 986443.
- M Linnerbauer, V Rothhammer. Protective Functions of Reactive Astrocytes Following Central Nervous System Insult. Front Immunol 11 (2020): 573256.


 Impact Factor: * 5.3
Impact Factor: * 5.3 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 