Modulatory Effect of Sodium Butyrate on the Anticancer Activity of Abemaciclib in MDA-MB-231 Human Breast Cancer Cells
Article Information
Neveen A. Elnozahi*, 1, Esraa A. Abdelaziz2, Maged W. Helmy3, Azza E. Bistawroos4
1Lecturer of pharmacology, Faculty of Pharmacy, Alexandria University, department of pharmacology and toxicology, Alexandria, Egypt
2Faculty of Pharmacy, Alexandria University, department of pharmacology and toxicology, Alexandria, Egypt
3Professor of pharmacology, Faculty of Pharmacy, Damanhour University, department of pharmacology and toxicology, Damanhour, Egypt
4Professor of pharmacology, Faculty of Pharmacy, Alexandria University, department of pharmacology and toxicology, Alexandria, Egypt
*Corresponding author: Neveen A. Elnozahi. Lecturer of pharmacology, Faculty of Pharmacy, Alexandria University, department of pharmacology and toxicology, Alexandria, Egypt
Received: 28 April 2023; Accepted: 05 May 2023; Published: 01 June 2023
Citation: Neveen A. Elnozahi, Esraa A. Abdelaziz, Maged W. Helmy, Azza E. Bistawroos. Modulatory effect of sodium butyrate on the anticancer activity of abemaciclib in MDA-MB-231 human breast cancer cells. Journal of Pharmacy and Pharmacology Research. 7 (2023): 62-73.
View / Download Pdf Share at FacebookAbstract
Triple negative breast cancer is the most aggressive subtype of breast cancer, and its treatment is limited. The effect of abemaciclib and /or sodium butyrate on MDA-MB-231 triple negative breast cancer cells was investigated. The IC50 for the growth inhibitory effects of abemaciclib, sodium butyrate, and their combination were 14.55 μM, 7.08 mM, and 3.743 mM, respectively. The combination showed a synergistic interaction with a decrease in IC50 to 2.55 μM abemaciclib and 3.74 mM butyrate. Three replicates of four groups of cancer cells were treated for 48 hr with the IC50 of abemaciclib, butyrate or their high or low dose combinations. A fifth group was treated with complete medium and served as the control. Cell migration, protein expression levels of cyclin D1, E2F2 transcription factor, phosphorylated AKT, nuclear factor kappa B (NF-κB), cyclin dependent kinase-2 (CDK2), retinoblastoma (Rb), p16INK4a, p53 and mRNA levels of CDK2, p16INK4a and p53 were assessed in all treated groups. Combination treatment with abemaciclib and butyrate showed a significant attenuation of cell metastasis. The combination treatment was associated with a decrease in E2F2, CDK2, and NF-κB protein levels together with attenuation in AKT phosphorylation level. The combination also showed an elevation in Rb, and p16INK4a, together with a reversal of DNA hypo-methylated state. Although abemaciclib monotherapy failed to alter cyclin D1 or p53 levels, the combination significantly reduced cyclin D1 level together with an increase in P53 level. In conclusion, combining butyrate with abemaciclib augmented its antiproliferative and antimetastatic effects and induced apoptotic activity.
Keywords
Breast cancer, TNBC, cell cycle, epigenetics, CDK4/6
Breast cancer articles, TNBC articles, cell cycle articles, epigenetics articles, CDK4/6 articles
Article Details
1. Introduction
Breast cancer is the second leading cause of death after lung cancer worldwide [1]. One in every 8 women has the risk of experiencing breast cancer throughout her life [2]. Breast cancer is classified according to histopathology of the cells, grade, stage and the molecular profile of the tumor [1]. According to the molecular profile, breast cancer is classified into four types: luminal A (estrogen receptor positive (ER+), progesterone receptor positive (PR+) and human epidermal growth factor receptor negative breast cancer (HER2-)), luminal B (ER+, PR+ and HER2+), HER2 enriched (ER-, PR- and HER2+), and triple negative breast cancer (TNBC), which lacks the expression of ER, PR, and HER2[3]. TNBC is one of the most aggressive subtypes of breast cancer, which is distinguished by a high proliferation rate and high ability to metastasize to distant organs such as the brain and lung, in comparison with other types that metastasize to the bone and soft tissues [4]. Due to the aggressiveness and lack of ER, PR and HER2 expression in this type of cancer, chemotherapy remains the only therapeutic option. In the last decade, several common hallmarks of cancer progression have been identified that participate in the transformation of normal cells into malignant cells. Among these hallmarks are genetic mutation or epigenetic alteration, uncontrolled cell proliferation, and resistance to apoptosis [5]. Epigenetic control of gene expression is defined as a mitotically heritable change in gene expression without changing DNA sequencing [6] via small tags on histone proteins or DNA sequences [7]. In human cells, the building unit of the chromosomes is the nucleosome, which consists of 147 base pairs of DNA wrapped around octamer histones (H2a, H2b, H3, and H4) and exterior H1 histone proteins that keep DNA wrapped in place. The structure of chromatin is controlled by acetylation and methylation of the lysine residue of the histone tails [7]. Posttranslational modifications of histones are controlled by histone acetyltransferases (HATs), histone deacetylases (HDACs), and histone methyltransferases (HMTs)[8]. The balance between these enzymes maintains the chromatin structure in an accurate state, leading to accurate gene expression according to cell needs and function [7]. In certain types of TNBC, there is an imbalance between those enzymes, such in mesenchymal stem- like breast cancer cells, which show an overexpression of HDACs [9]. HDACs are classified into 4 classes according to their structure: class I (HDAC 1,2,3,8), class IIa (HDAC 4,5,7,9)/class IIb (HDAC 6,10), class III (SIRT1-7), and class IV (HDAC 11). These enzymes have the ability to remove acetyl groups from histone proteins, thereby affecting gene expression and removing acetyl groups from expressed nonhistone proteins, thus altering their biological activity [10].
DNA methylation is the second most common epigenetic modification; methylation of cytosine-guanine (CpG) islands at the gene promoter leads to silencing of gene expression, while methylation of the gene body leads to activation of gene expression. Alterations in DNA methylation in cancer are common, as most cancer types show global hypomethylation at CpG islands with specific hypermethylation at the promoter of tumor suppressor genes, leading to genetic instability and silencing of tumor suppressors [11]. As epigenetic alterations have a remarkable contribution to tumor formation, most recent studies have focused on epigenetic drugs as antitumor therapies [12]. One of the most common epigenetic drugs is HDAC inhibitors, such as sodium butyrate (NaBu) [13]. NaBu is a short chain fatty acid that is produced naturally in the colon due to bacterial fermentation of fibers, and it acts as an inhibitor of HDAC class I/IIa/IV [14]. The antitumor activity of NaBu was reported in previous 3 studies against various types of cancer, including breast cancer [15,16]. The cell cycle process is controlled by cyclins and cyclin dependent kinases, which are defined as cell cycle drivers. The cell cycle is also governed by cyclin dependent kinase inhibitors, which are cell cycle brakes. Uncontrolled cell proliferation is caused by alterations in cell cycle regulator protein and gene expression [17]. In the last decade, synthetic drugs that target the cell cycle pathway have been developed such as palbociclib, ribociclib, and abemaciclib. These drugs are approved by the FDA for treating metastatic hormonal positive breast cancer, as they improve patient outcomes and significantly increase the survival rate [18]. Abemaciclib is an ATP competitive inhibitor of CDK4/6 with a higher selectivity for CDK4 over CDK6. This reduces the degree of neutropenia usually associated with inhibition of CDK6, thus avoiding the need for an interruption in dosing as with other CDK4/6 inhibitors. Abemaciclib was approved on February 2018 by the FDA for the treatment of advanced metastatic hormonal positive breast cancer as a monotherapy or in combination with endocrine therapy [18].
Despite the success of abemaciclib in breast cancer treatment, some patients showed acquired or denovo resistance. Several mechanisms attribute to the development of abemaciclib resistance such as the elevation of CDK2 expression [19]. Abemaciclib monotherapy also showed an accumulation in cyclin D1 in colon cancer cells which attenuated its anti-proliferative activity [2]. NaBu significantly decreased cyclin D1 in many types of cancer such as colon cancer [15,21,22]. NaBu also decreased CDK2 in monoplastic leukemia cells (M. Huang et al., 2004). This prompted us to investigate the possible modulatory effect of NaBu on the antiproliferative activity of abemaciclib. The study also aimed to investigate the effect of this combination on some of the multiple mechanisms that contribute to abemaciclib resistance.
2. Material and Methods
2.1 Cell culture
Human MDA-MB-231 triple negative breast cancer cells were purchased from American Type Culture Collection (ATCC) and conducted in the Center of excellence for Research in Regenerative Medicine and its Applications (CERRMA). Breast cancer cells were cultured in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% penicillin-streptomycin under a humidified atmosphere containing 5% CO2 at 37°C. The cells (70% confluent) were treated with 14.5 μM abemaciclib LY2835219 (Selleckchem Co-USA) and/or 7 mM NaBu (Sigma Co-USA) and incubated in aCO2 incubator for 48 h.
2.2 Cell viability assay
To investigate the growth inhibitory effect of abemaciclib and NaBu as single drugs or in combination, MDA-MB-231 cells were transferred to 96-well plates at a density of 7000 cells/well and incubated at 37°C for 24 h. The cells were then treated with different concentrations of abemaciclib (0.09-11.58 μM) and/or different concentrations of NaBu (1.325-16.96 mM) and incubated for an additional 48 h. The effect of all single and combinatorial treatments on cell viability was assessed using a methylthiazolydiphenyl-tetrazolium bromide (MTT) assay [23]. Briefly, after the incubation period, MTT reagent was added to each well at a final concentration of 0.5 mg/ml. The plate was incubated at 37°C for 4 h in a CO2 incubator. Then, 100 μL of dimethyl sulfoxide (DMSO) was added to solubilize formazan crystals. Then the plate was shaken gently in darkness for 15 min. The optical density (OD) was measured at 590 nm using an ELISA microplate reader. The relative viability of each treatment was analyzed based on the absorbance of each treated sample compared to the control sample. The IC50 of each single drug and their combination were determined using CompuSyn software. To determine whether the interaction was synergistic or antagonistic , the combination index (CI) was determined as described by Chou [24], where CI<1 indicates synergism, =1 indicates additive action, and >1 indicates antagonism. The dose reduction index (DRI), which signifies how many folds of dose reduction for each drug at a given effect are allowed in synergistic combination, was then determined. Five experimental groups were used. The first group was treated with complete medium and served as a control group. The four remaining groups were treated with abemaciclib, NaBu or their combinations for 48 hr. Three replicates of each group were treated with the IC50 of each single drug or their combination for 48 hr. The nuclear extract or cell lysate was processed by RT-PCR or ELISA, respectively to quantify the mRNA level and protein level of the investigated parameters.
2.3 Quantitative real-time PCR
Total RNA was extracted using a miRNeasy mini kit (#217004, QIAGEN). Then, total mRNA was reverse transcribed to complement DNA (cDNA), and quantitative real-time polymerase chain reaction (qRT-PCR) was performed using a Rotor-Gene SYBR Green Kit (#204174, QIAGEN) according to the manufacturer’s instructions. Quantitative RT-PCR was performed using a Rotor-Gene cycler Q-pure detection system version 2.1.0 (QIAGEN, USA). The primers used in qRT-PCR analysis were as follows: human TP53 (5’-CCAGCCAAAGAAGAAACCA -3’ Forward, 5’- TATGGCGGGAGGTAGACTGA-3’ Reverse); human CDKN2A (5′-CCCAACGCACCGAATAGTTA-3′ Forward, 5′-GCATGGTTACTGCCTCTGGT-3′ Reverse); human CDK2 (5′-GTCCCCAGAGTCCGAAAGAT-3′ Forward, 5′-GCTTTCTGCCATTCTCATCG-3′ Reverse). Human β-actin was chosen as an endogenous control (housekeeping gene). For one step RT-PCR, the reaction mixture (25 μl) contained 1 μl template RNA, 12.5 μl 2x Rotor-gene SYBR Green RT-PCR Master Mix, 2.5 μl of each set of oligonucleotide primers, 0.25 μl Rotor-Gene RT Mix, and 6.25 μl RNase-free water. The one step real-time PCR conditions included a reverse transcription at 55°C for 10 min, an initial denaturation at 95°C for 5 min, followed by a 40-cycle amplification consisting of denaturation at 95°C for 5 sec, and combined annealing/extension at 60°C for 10 sec followed by melting curves to verify qRT-PCR product identity. All reactions were examined in triplicate for each sample. All primer pairs were checked using primer blast. Relative mRNA expression levels were calculated by the 2-ΔΔCt method.
2.4 Enzyme-linked immunosorbent assay (ELISA)
Quantification of the protein expression level of cell cycle regulatory proteins in control and treated cells was measured using ELISA Kits. The proteins detected were human cyclinD1 (ab214571 SimpleStep, Abcam), human PRb using human PRb (MBS2508443, Mybiosources), E2F transcription factor 2 (E2F2) (#abx151366, Abbexa,UK), human CDK2 using human CDK2 (#MBS2510990, Mybiosources), CDKN2A (p16) (ab227903 SimpleStep, Abcam), p53 using (ab171571-p53 human SimpleStep, Abcam), phosphorylated AKT (pS473) (#KHO0111, Invitrogen), and NF-κB (#MBS450580, Mybiosources). Global DNA methylation (5-methylcytosine) was determined using an ab233486 colorimetric kit (Abcam). All kits were used according to the manufacturer’s protocol.
2.5 Wound healing assay
MDA-MB-231 cells were seeded into 6 well plates and left to grow for 48 h. Confluent monolayers were gently scratched using a sterile 1 ml pipette tip across the center of the well. The cells were washed twice with complete media to remove detached cells, and then the cells were replenished with 2 ml of new complete media supplemented with 10% FBS and 1% Pen-Strep. Afterward, abemaciclib and/or NaBu were added to the wells. Images were captured for each well at zero time and after incubation for 24 h and 48 h using an inverted microscope. The percent cell migration was expressed as the number of migrated treated cells relative to the control [25]. The following equation was used for its
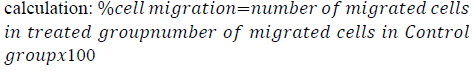
2.6 Statistical analysis
All results are presented as the mean ± S.D. The computer software GraphPad Prism version 9.00 for Windows, GraphPad Software, USA, was used for all statistical analyses and graphical data presentations. For the comparison between experimental groups, one-way analysis of variance (ANOVA) (nonparametric) was used, followed by Tukey’s multiple comparisons test. Differences with values of P<0.05 were considered statistically significant.
3. Results
3.1 Cytotoxicity assay
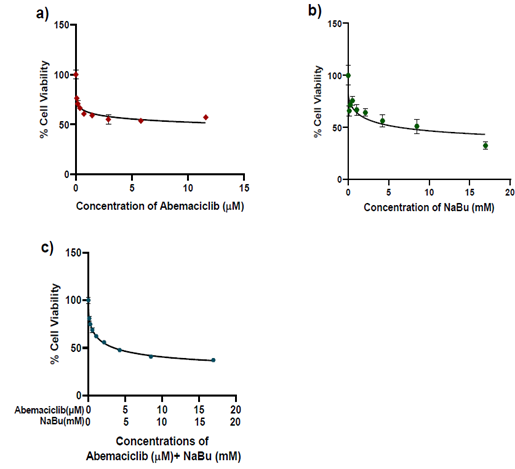
The growth inhibitory effect of abemaciclib and NaBu (alone or in combination) on MDA-MB-231 cells was determined using the MTT assay. The viability of MDA-MB-231 cells was reduced after abemaciclib and/or NaBu treatment for 48 h in a concentration dependent manner (Fig.1). The IC50 values of abemaciclib, NaBu, and their combination were 14.55 μM, 7.08 mM, and 3.743 mM, respectively. Microscopic examination confirmed the higher potency of the combination on cell viability (Fig. 2). The combination of abemaciclib and NaBu showed a synergistic interaction, as the calculated CI value was found to be less than 1 (CI = 0.70399). The calculated DRIs of both drugs at 50% cell viability were 5.69 and 1.89 for abemaciclib and NaBu, respectively, and it was translated into a decrease in IC50 equal to 2.55 μM abemaciclib and 3.74 mM NaBu. Three replicates of four groups of cancer cells were treated for 48 hr with the IC50 of abemaciclib, butyrate or their combinations. Two combinations were used: high-dose combination (combination 1) and low-dose combination according to the dose reduction index (combination DRI). A fifth group treated with complete medium served as the control, as shown in Table (1).
Table 1: Experimental groups, and the IC50 of each drug in treated groups
|
Experimental group |
IC50 of abemaciclib |
IC50 of NaBu |
|
Control |
- |
- |
|
Abemaciclib |
14.55μM |
- |
|
NaBu |
- |
7.08 mM |
|
Combination1 |
14.55μM |
7.08 mM |
|
Combination (DRI) |
2.55μM |
3.74 mM |
3.2 Expression of the cell cycle drivers cyclinD1, E2F2, and CDK2
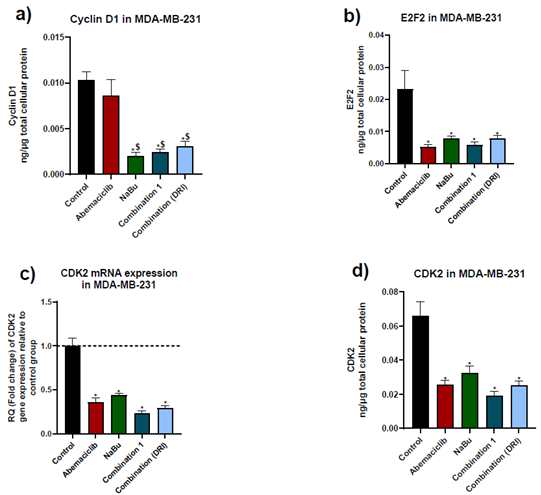
Abemaciclib as a single treatment caused a remarkable decrease in E2F2 protein level and CDK2 gene and protein levels in comparison with untreated cells, without showing any significant change in the protein expression level of cyclin D1. On the other hand, NaBu as a monotherapy showed a significant decrease in cyclin D1 and E2F2 protein levels, and CDK2 gene and protein levels. Combinations treatment showed a significant decrease in cyclin D1 protein level in comparison with abemaciclib as a single drug (Fig. 3).
Figure 3: Effects of 48 hr treatment of MDA-MB-231 human breast cancer cell line with abemaciclib, NaBu, or their combinations on; a) cyclinD1 protein expression level, b) E2F2 protein expression level, c) cyclin dependent kinase 2 (CDK2) mRNA gene expression level and d) CDK2 protein expression level. Data are expressed as the mean ± SD (Standard deviation) of three samples each performed in triplicate. * P< 0.05 vs control, $ P<0.05 vs abemaciclib
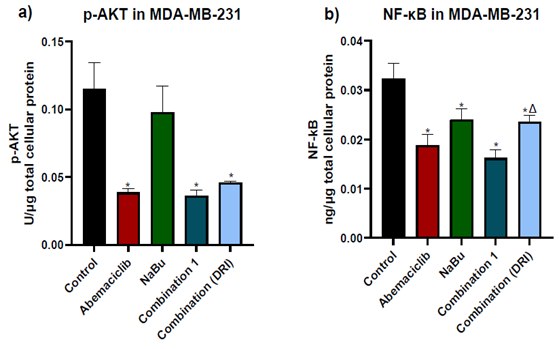
3.3 AKT phosphorylation level and NF-κB protein expression level.
The phosphorylation level of AKT at S473 and the NF-κB protein level were determined in all experimental groups. As shown in figure (4), abemaciclib as a single drug caused a remarkable decrease in both the AKT phosphorylation level at S473 and the NF-κB protein expression level in comparison with untreated cells. On the other hand, NaBu as a single drug significantly reduced NF-κB protein expression level and had no effect on AKT phosphorylation level at S473 in comparison with untreated cells (control). The combinations treatment did not further decrease neither in AKT phosphorylation level or NF-κB protein expression level compared with abemaciclib as a single drug
Figure 4: Effects of 48 hr treatment of MDA- MB-231 human breast cancer cells with abemaciclib, NaBu, or their combinations on; a) AKT phosphorylation level at S473 and b) NF-κB protein expression level. Data are expressed as the mean ± SD (Standard deviation) of three samples each performed in triplicate. *P< 0.05 vs control, $ P< 0.05 vs abemaciclib, Δ P< 0.05 vs combination1
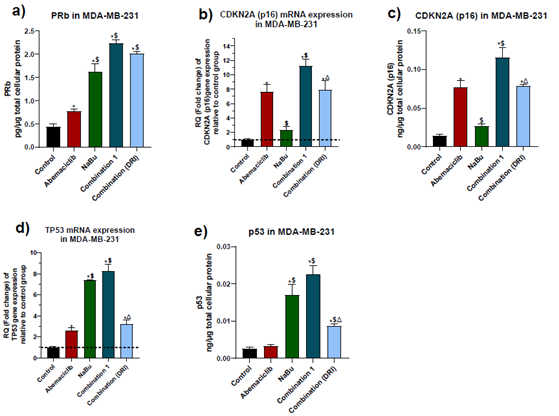
3.4 Expression levels of the tumor suppressors PRb, p16 and p53.
The protein level of the hypophosphorylated form of retinoblastoma (Rb) and the gene and protein expression levels of p16 and wild-type p53 tumor suppressors were measured. As shown in figure (5), abemaciclib caused a significant elevation in PRB protein level, p16 gene expression and protein levels, but its upregulating effect on p53 was restricted to gene expression only without any significant elevation in protein level, in comparison with the untreated cells. On the other hand, NaBu showed a significant elevation in PRb protein level, p53 gene expression and p53 protein levels, but it had no effect on p16 gene or protein expression levels. The combinations treatment showed a further elevation in PRB protein expression level in comparison with each single drug. While combination 1 produced a significant elevation in p16 gene and protein expression levels and p53 protein level as compared with abemaciclib or NaBu, the p53 gene expression level was elevated in comparison with abemaciclib only. Compared with abemaciclib, combination 2 significantly elevated p53 protein expression level without significantly altering P53 gene level
Figure 5: Effect of 48 hr treatment of MDA-MB-231 human breast cancer cell line with abemaciclib, NaBu, or their combinations on: a) PRb protein expression level, b) p16 gene expression level, c) p16 protein expression level, d) p53 gene expression level, e) p53 protein expression level. Data are expressed as the mean ± SD of three samples each performed in triplicate. *P< 0.05 vs control, $ P< 0.05 vs abemaciclib, Δ P< 0.05 vs combination1
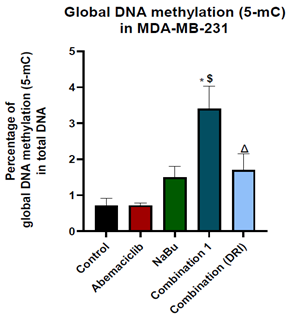
Figure 6: Effect of 48 hr treatment of MDA-MB-231 human breast cancer cell line with abemaciclib, NaBu, or their combinations on global DNA methylation at 5-methyl cytosine. Data are expressed as the mean ± SD of three samples each performed in triplicate. *P< 0.05 vs control, $ P< 0.05 vs abemaciclib, Δ P< 0.05 vs combination1
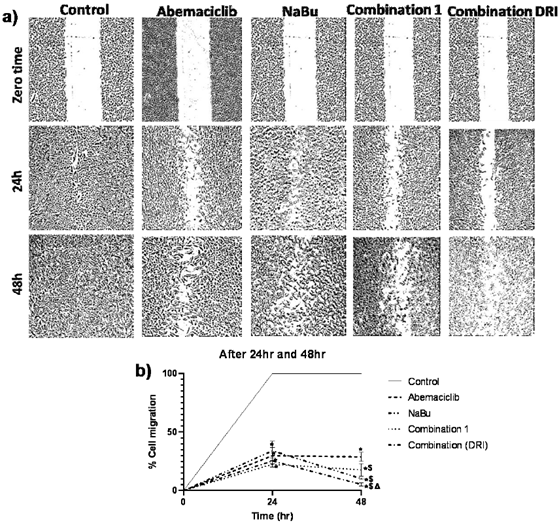
3.5 Wound healing assay
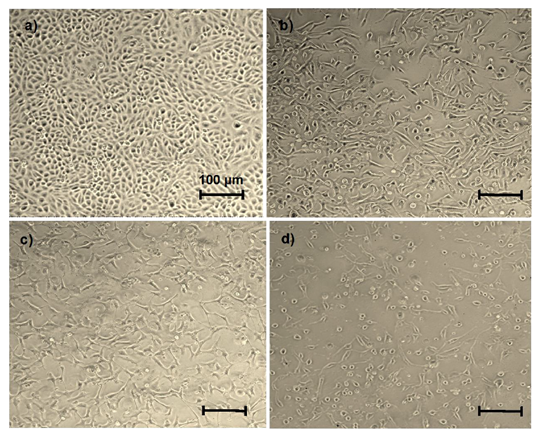
To investigate the effect of abemaciclib and NaBu (alone or in combination) on cell migration in MDA-MB-231cells, the percent reduction in cell migration was assessed by the wound healing assay. As shown in figure (7), after 24 hr or 48 hr of treatment with each single drug alone or in combination the cell migration was significantly reduced in comparison with the untreated cells. Compared to abemaciclib, NaBu and combinations treatment significantly reduced cell migration after 48 hr. The inhibitory effect of combination 2 (DRI) on cell migration was significantly higher than that of combination 1 after 48 hr.
Figure 7: Effect of treatment of the MDA-MB-231 human breast cancer cell line with abemaciclib, NaBu, or their combinations on cell migration. A) Microscopic examination of cell migration at zero time, 24 hr and 48 hr. B) Line graph showing % cell migration at zero time and after 24 hr and 48 hr. Data are expressed as the mean ± SD (Standard deviation) of three samples each performed in triplicate. *P<0.05 vs control, $ P< 0.05 vs abemaciclib, Δ P< 0.05 vs combination1
4. Discussion
Disturbances in HDAC expression is implicated as a possible carcinogenesis trigger, due to their vital role in controlling the expression of many oncogenes and tumor suppressor genes as well as their involvement in controlling the activity of several transcriptional factors [10]. Uncontrolled cell proliferation is one of the hallmarks of cancer, and one of its triggers might be through nuclear and cytoplasmic disturbances in HDAC expression [7]. The reported alteration in HDAC expression in MDA-MB-231 cells [9] prompted us to investigate the possible modulatory effect of NaBu on abemaciclib anticancer activity.
In the present study, MDA-MB-231 cells were found to overexpress cyclin D1, and upon treating the cells with abemaciclib for 48 h, cyclin D1 protein level was not reduced. This result is consistent with a previous study in which abemaciclib failed to decrease cyclin D1 expression in colorectal cancer [20]. Abemaciclib also showed an elevation in the hypophosphorylated form of Rb tumor suppressor protein in the present study. Consistently, it was reported that abemaciclib treatment caused the accumulation of cyclin D1 together with a partial inhibition of Rb phosphorylation in the HCT116 colorectal cancer cell line [26]. It is well recognized that abemaciclib, through its inhibitory effect on CDK4/6, results in the inhibition of Rb phosphorylation which restricts cell cycle progression [18]. To confirm the implication of cyclin D1 accumulation in the partial inhibition of Rb phosphorylation, cyclin D1 knockout cells were treated with abemaciclib, and complete inhibition of Rb phosphorylation was noted in colorectal cancer cells [26].
In the present study, NaBu was able to reduce cyclin D1 protein level and elevate the hypophosphorylated form of Rb. This is consistent with the previously reported decrease in cyclin D1 level upon treatment of a monoplastic leukemia cell line (SKM-1) and a nonsmall lung cancer cell line (NCI-H23) with NaBu [27,28]. The ability of NaBu to decrease cyclin D1 expression might be due to its inhibitory effect on NF-κB, which is considered a direct transcriptional activator of cyclin D1 [21]. NaBu could also inhibit cyclin D1 expression through inhibition of the cytoplasmic HDAC6 [9]. Briefly, inhibition of HDAC6, leads to hyperacetylation and proteosomal degradation of HSP90, which in turn leads to destabilization and degradation of EGFR, thus attenuating all mitogenic signaling pathways downstream of EGFR activation, such as the MAPK/ERK1-2 and PI3K/AKT/mTOR signaling pathways [29]. Inhibition of the MAPK/ERK1-2 pathway attenuates the formation of the AP-1 transcription factor, which is a direct activator of cyclin D1 expression. In addition, inhibition of the PI3K/AKT/mTOR signaling pathway activates GSK-3β and leads to phosphorylation and proteosomal degradation of cyclin D1 protein [29-31]. Various possible mechanisms could be linked to the effect of NaBu on Rb phosphorylation. First, NaBu is able to induce wild type p53 expression [15], which was confirmed in the present study, as NaBu elevated both the gene and protein levels of p53. As p53 is a stress detector transcription factor, its expression is elevated during DNA damage and nutrient deficiency or any sort of cellular stress, such as that induced by drugs or free radicals. The expression of p53 controls cell fate, as one of its roles is to directly induce the transcription of p21 and p27, which are endogenous CDK/cyclin complex inhibitors, with a consequent attenuation of Rb phosphorylation level, to stop cell cycle progression and give much more time to the cell to repair DNA damage or to restore energy in the case of nutrient deficiency [17]. Thus the ability of NaBu to induce p53 expression could be linked to its ability to induce cell cycle arrest, as was noted in HCT116 colon cancer cells treated with NaBu [32].
It is worth mentioning that NaBu could activate p21 and p27 transcription through inhibition of the PI3K/AKT signaling pathway secondary to inhibition of HDAC6 as mentioned before [29,30]. Another possible mechanism for the diminished Rb phosphorylation by NaBu treatment could be mediated through the inhibition of nuclear HDAC1, which is an inhibitor of the SP-1 transcription factor. Activation of SP-1 could also induce the transcription of p21 [33]. In addition, NaBu was reported to decrease c-myc oncogene expression which acts as a transcriptional repressor of p21 [15,34]. In the present study the combination of NaBu and abemaciclib enhanced the ability of abemaciclib to induce cell cycle arrest by decreasing cyclin D1 expression with a further elevation in hypophosphorylated Rb level and the effect was maintained in a small dose combination. Abemaciclib and NaBu as single drugs were able to reduce the expression of the cell cycle driver E2F2 in the current study. This decrease could be mediated through their ability to reduce c-myc expression which is a direct activator of E2F2 expression [35]. Previously, a significant decline in c-myc level upon treatment of mutant non-small lung cancer cells with abemaciclib or NaBu treatment was noted [34]. It was also reported that the degradation of HSP90 induced by NaBu causes destabilization and degradation of E2F1,E2F2 and E2F3 [36].
Abemaciclib and NaBu as single drugs, attenuated CDK2 gene and protein levels. Consistent with this finding, it was reported that abemaciclib reduced CDK2 expression in various cancer cells and was considered a pharmacodynamic biomarker, indicating the responsiveness of the patient to the treatment [18]. A significant decrease in CDK2 expression upon NaBu treatment was also previously noted in monoplastic leukemia cells [27]. This decrease could be due to the observed decrease in the NF-κB level, which acts as a direct transcriptional activator of the CDK2 gene, as well as an indirect activator by increasing the E2F3 level [37].
In the present study, AKT phosphorylation level and NF-κB protein expression level were analyzed as both are considered mitogenic signaling nodes implicated in tumorigenesis process. NF-κB is also a direct down-streamer to AKT phosphorylation [30]. In addition, PI3K/AKT signaling pathway is considered a non-cell cycle specific mechanism attributing to abemaciclib resistance [19]. Moreover, NF-κB overexpression was observed in MDA-MB-231 cells and was implicated in tumor progression [38].
Recently, a link between CDK6 inhibition and NF-κB phosphorylation, particularly the P65 subunit, was noted in cervical cancer cell lines. CDK6 phosphorylates the p65 subunit and eases its translocation to the nucleus, leading to the transcription of genes implicated in cell proliferation, angiogenesis, metastasis and apoptosis resistance [39]. Thus, inhibition of CDK6 by abemaciclib may lead to attenuation of NF-κB transcriptional activity and its expression.
On the other hand, acetylation of the p65 subunit could activate or repress p65 transcriptional activity according to the site of the acetyl group. HDAC3 has the ability to remove the acetyl group from inhibitory residues on the p65 subunit. Thus, inhibition of HDAC3 by NaBu could attenuate p65 transcriptional activity and in turn decrease NF-κB level [40]. Abemaciclib reduced AKT phosphorylation level in the present study which is in parallel with the abemaciclib-induced decrease in the non-small lung cancer cell line [34]. In contrast to other CDK4/6 inhibitors, such as palbociclib, which showed an elevation in AKT phosphorylation level in the MDA-MB-231 breast cancer cell line. On the other hand, NaBu had no effect on AKT phosphorylation as a single drug despite its ability to attenuate the EGFR signaling pathway [16]. A similar observation was noted in a cervical cancer cell line [32].
By analyzing the cell cycle regulators, abemaciclib significantly elevated p53 gene expression but failed to stabilize its protein level. Similarly, abemaciclib failed to induce p53 level in a liposarcoma cell line [41]. In various cancer types, abemaciclib failed to induce apoptosis and its effect was restricted to the inhibition of cell proliferation [20,34]. On the other hand, NaBu showed a significant elevation in p53 gene and protein levels. This finding is in line with the reported induction of intrinsic apoptosis, through p53-dependent pathways, in the MCF7 breast cancer cell line and HCT116 colon cancer cell line following butyrate treatment [15]. The mechanism by which NaBu stabilizes p53 protein level might be through its inhibitory effect on HDAC1. Normally, HDAC1 deacetylates p53 protein leading to recruitment of MDM2 E3 ubiquitin ligase, and proteosomal degradation of p53[10]. Stabilization of p53 by NaBu allows it to binds to proapoptotic promoters and enhances their transcription [15]. It is worth mentioning that, the combination treatment showed a highly significant elevation in the p53 gene more than each drug alone. It could be suggested that, despite the ability of abemaciclib to reduce AKT phosphorylation, and NF-κB protein level which are correlated with the inhibition of MDM2 activity and expression respectively, it failed to stabilize p53 protein. Upon adding NaBu in combination with abemaciclib, p53 stabilization was augmented through inhibition of MDM2 recruitment facilitated by inhibition of HDAC1 activity. This combination would be highly beneficial in MDA-MB-231 cancer cells, as these cells are reported to overexpress both MDM2 [42] and HDAC1[43]. Stabilization of p53 was observed even with the low dose combination; thus the lower dose combination could be recommended to maintain the cytocidal effect while preventing the large dose side effects.
The effect of each single drug or the combinations on p16 was investigated. p16 is an endogenous CDK4/6 inhibitor that regulates cell cycle progression in normal cells as it acts as a cell cycle brake[17]. Cancer cells rely on attenuating those cell cycle brakes to ease the cell proliferation process during tumorigenesis [17]. It was reported that MDA-MB-231 cells show a low expression level of p16 [44]. On the other hand, overexpression of p16 could be implicated in tumorigenesis, by inducing genomic instability, which might lead to additional mutations, such as KRAS mutations. Overexpression of p16 may also lead to a decrease in cell dependence on CDK4/6 and overexpression of cyclin E and CDK2 to complete cell cycle progression [45]. In addition, overexpressed p16 may translocate to the cytoplasm and enhance oncogenic biological functions[45,46]. Therefore, p16 overexpression is considered one of the cell cycle specific mechanisms of resistance against CDK4/6 inhibitors[47]. In the present study abemaciclib caused a significant elevation in p16 gene and protein expression levels, On the other hand NaBu as a single drug had no effect on p16 expression level. Similarly, Pellizzaro,et al.,reported that NaBu caused no change in p16 expression level in the NCI-H23 non-small cell lung cancer cell line[28]. However, in colon cancer cells, butyrate caused an elevation in p16 expression [22].
Combination treatment with NaBu and abemaciclib showed a significant increase in p16 gene and protein levels in comparison with abemaciclib. This effect was evident only with the high dose combination. Thus using the low dose combination could be preferable as the high dose may enhance abemaciclib resistance by increasing p16 level. One of the characteristic features of TNBC is its high ability to migrate to distant organs, especially the brain and lung, which contributes to making TNBC a lethal and aggressive breast cancer subtype. The molecular mechanism that underlies cell migration in TNBC is the low cadherin 3/4, and the high level of EMT biomarkers such as snail [48]. In the present study, after 24 h of treatment, the cell migration was significantly reduced in all treated groups with no differences between the single drugs and the combination treatment. After 48 h, NaBu and the combination treatment showed a further decrease in cell migration in comparison with abemaciclib. The antimigration effect was more pronounced in the low dose combination, supporting the beneficial value of the low dose combination over the high dose combination. Antimetastatic effects of abemaciclib and NaBu were previously noted in Caco-2 colorectal cancer cells [20] and MDA-MB-231 cells respectively [43].
The antimigration effect of both drugs may be linked to their ability to reduce NF-κB and elevate active RB protein levels. NF-κB acts as a direct transcription activator of metalloproteinase 9 (MMP-9)[49]. MMP-9 has the ability to degrade the extracellular matrix facilitating the migration of cells to the bloodstream and/or lymph nodes [49]. NF-κB also stabilizes snail protein [50], which acts as a direct transcriptional repressor of the cell adhesion molecule E-cadherin. Additionally, Rb protein acts as a direct transcriptional activator of E-cadherin [51]. Butyrate exerts an additional mechanism, as it attenuates AP-1 complex formation via inhibition of MAPK/ERK1-2[16]. The AP-1 complex is implicated in metastasis as it is a transcription factor that induces the expression of MMP-9. [49]. In conclusion, as TNBC is the most aggressive breast cancer subtype, the need to develop a novel therapeutic protocol is of great importance. Combining NaBu with abemaciclib could be a new therapeutic avenue as it enhanced the antitumor activity of abemaciclib through epigenetic regulation of cyclin D1, Rb, and p53 expression levels. The combination also showed an augmented antimetastatic effect, which was more pronounced with low-dose treatment. This combination could improve treatment outcome and patient survival, as well as achieving the most challenging issue in cancer treatment, that is, reducing the dose without losing drug effectiveness.
Statements and declarations
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Competing Interests
The authors have no relevant financial or nonfinancial interests to disclose.
Author Contributions
The authors declare that all data were generated in-house and that no paper mill was used. All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Esraa A. Abdelaziz. The first draft of the manuscript was written by Esraa A. Abdelaziz and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval:
Not applicable
Consent to participate:
Not applicable
Consent for publication:
Not applicable
Code availability:
Not applicable
References
- Rakha EA, Reis-Filho JS, Baehner F, Dabbs DJ, Decker T, Eusebi V., . . . et al. Breast cancer prognostic classification in the molecular era: the role of histological grade. Breast Cancer Res, 12 (2010): 207.
- Kaminska M, Ciszewski T, Lopacka-Szatan K, Miotla P, & Staroslawska, E. Breast cancer risk factors. Prz Menopauzalny, 14 (2015): 196-202.
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA., . . . et al. Molecular portraits of human breast tumours. Nature, 406 (2000): 747-752.
- Uscanga-Perales GI, Santuario-Facio SK & Ortiz-López R. Triple negative breast cancer: Deciphering the biology and heterogeneity. Medicina Universitaria 18 (2016): 105-114.
- Hanahan D & Weinberg RA. Hallmarks of cancer: the next generation. Cell 144 (2011): 646-674.
- Jovanovic J, Rønneberg JA, Tost J, & Kristensen V. The epigenetics of breast cancer. Mol Oncol 4 (2010): 242-254.
- Sharma S, Kelly TK, & Jones PA. Epigenetics in cancer. Carcinogenesis 31 (2010): 27-36.
- Black J C, Van Rechem C & Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 48 (2012): 491-507.
- Kalle AM & Wang Z. Differential effects of two HDAC inhibitors with distinct concomitant DNA hypermethylation or hypomethylation in breast cancer cells (2019): 578062.
- Seto E & Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol 6 (2014): a018713.
- Kondo Y. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med J 50 (2009): 455-463.
- Temian DC, Pop LA, Irimie AI & Berindan-Neagoe I. The Epigenetics of Triple-Negative and Basal-Like Breast Cancer: Current Knowledge. J Breast Cancer, 21 (2018): 233-243.
- Garmpis N, Damaskos C, Garmpi A, Kalampokas E, Kalampokas T, Spartalis E., . . . et al.. Histone Deacetylases as New Therapeutic Targets in Triple-negative Breast Cancer: Progress and Promises. Cancer Genomics Proteomics 14 (2017): 299-313.
- Damaskos C, Garmpis N, Valsami S, Kontos M, Spartalis E, Kalampokas T., . . . et al. Histone Deacetylase Inhibitors: An Attractive Therapeutic Strategy Against Breast Cancer. Anticancer Res 37 (2017): 35-46.
- Chen J, Zhao KN, & Vitetta L. Effects of Intestinal Microbial Elaborated Butyrate on Oncogenic Signaling Pathways. Nutrients 11 (2019).
- Huang W, Zeng C, Liu J, Yuan L, Liu W, Wang L., . . . et al. Sodium butyrate induces autophagic apoptosis of nasopharyngeal carcinoma cells by inhibiting AKT/mTOR signaling. Biochem Biophys Res Commun, 514 (2019): 64-70.
- Vermeulen K, Van Bockstaele DR, & Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif, 36 (2003): 131-149.
- O'Brien N, Conklin D, Beckmann R, Luo T, Chau K, Thomas J, . . . et al. Preclinical Activity of Abemaciclib Alone or in Combination with Antimitotic and Targeted Therapies in Breast Cancer. Mol Cancer Ther 17 (2018): 897-907.
- Guarducci C, Bonechi M, Boccalini G, Benelli M, Risi E, Di Leo A., . . . et al. Mechanisms of Resistance to CDK4/6 Inhibitors in Breast Cancer and Potential Biomarkers of Response. Breast Care (Basel) 12 (2017): 304-308.
- Lee HJ, Kim KJ, Sung JH, Nam M, Suh KJ, Kim JW., . . . et al. PI3K p110α Blockade Enhances Anti-Tumor Efficacy of Abemaciclib in Human Colorectal Cancer Cells. Cancers (Basel) 12 (2020).
- Hu J, & Colburn NH. Histone deacetylase inhibition down-regulates cyclin D1 transcription by inhibiting nuclear factor-kappaB/p65 DNA binding. Mol Cancer Res 3 (2005): 100-109.
- Schwartz B, Avivi-Green C & Polak-Charcon S. Sodium butyrate induces retinoblastoma protein dephosphorylation, p16 expression and growth arrest of colon cancer cells. Mol Cell Biochem 188 (1998): 21-30.
- van Meerloo J, Kaspers GJ & Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol Biol 731 (2011): 237-245.
- Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res 70 (2010): 440-446.
- Xiao X, He Z, Cao W, Cai F, Zhang L, Huang Q, . . . Liu, Y. Oridonin inhibits gefitinib-resistant lung cancer cells by suppressing EGFR/ERK/MMP-12 and CIP2A/Akt signaling pathways. Int J Oncol 48 (2016): 2608-2618.
- Chen SH, Gong X, Zhang Y, Van Horn RD, Yin T, Huber L, . . . et al. RAF inhibitor LY3009120 sensitizes RAS or BRAF mutant cancer to CDK4/6 inhibition by abemaciclib via superior inhibition of phospho-RB and suppression of cyclin D1. Oncogene 37 (2018): 821-832.
- Huang M, Liu W, Li C, Deng J, Zhou J, Zhang D, & Sun H. The effect of sodium butyrate in combination with ATRA on the proliferation/differentiation of SKM-1. J Huazhong Univ Sci Technolog Med Sci 24 (2004): 334-337.
- Pellizzaro C, Coradini D, Daniotti A, Abolafio G, & Daidone MG. Modulation of cell cycle-related protein expression by sodium butyrate in human non-small cell lung cancer cell lines. Int J Cancer 91 (2001): 654-657.
- Wee P, & Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel) 9 (2017).
- Ghoneum A, & Said N. PI3K-AKT-mTOR and NFκB Pathways in Ovarian Cancer: Implications for Targeted Therapeutics. Cancers (Basel) 11 (2019).
- Guo ZY, Hao XH, Tan FF, Pei X, Shang LM, Jiang XL, & Yang F. The elements of human cyclin D1 promoter and regulation involved. Clin Epigenetics 2 (2011): 63-76.
- Chen J, Ghazawi FM, Bakkar W, & Li Q. Valproic acid and butyrate induce apoptosis in human cancer cells through inhibition of gene expression of Akt/protein kinase B. Mol Cancer 5 (2006): 71.
- Pajak B, Orzechowski A, & Gajkowska B. Molecular basis of sodium butyrate-dependent proapoptotic activity in cancer cells. Adv Med Sci 52 (2007): 83-88.
- La Monica S, Fumarola C, Cretella D, Bonelli M, Minari R, Cavazzoni A, . . . et al. Efficacy of the CDK4/6 Dual Inhibitor Abemaciclib in EGFR-Mutated NSCLC Cell Lines with Different Resistance Mechanisms to Osimertinib. Cancers (Basel) 13 (2020).
- Beier R, Bürgin A, Kiermaier A, Fero M, Karsunky H, Saffrich R, . . . et al. Induction of cyclin E-cdk2 kinase activity, E2F-dependent transcription and cell growth by Myc are genetically separable events. Embo j 19 (2000): 5813-5823.
- Kotwal A, Suran S, & Amere Subbarao S. Hsp90 chaperone facilitates E2F1/2-dependent gene transcription in human breast cancer cells. Eur J Cell Biol 100 (2021): 151148.
- Ledoux AC, & Perkins ND. NF-κB and the cell cycle. Biochem Soc Trans 42 (2014): 76-81.
- Liu B, Wen JK, Li BH, Fang XM, Wang JJ, Zhang YP, . . . et al. Celecoxib and acetylbritannilactone interact synergistically to suppress breast cancer cell growth via COX-2-dependent and -independent mechanisms. Cell Death Dis, 2 (2011): e185.
- Buss H, Handschick K, Jurrmann N, Pekkonen P, Beuerlein K, Müller H, . . . et al. Cyclin-dependent kinase 6 phosphorylates NF-κB P65 at serine 536 and contributes to the regulation of inflammatory gene expression. PLoS One, 7 (2012): e51847.
- Leus NG, Zwinderman MR, & Dekker FJ. Histone deacetylase 3 (HDAC 3) as emerging drug target in NF-κB-mediated inflammation. Curr Opin Chem Biol, 33 (2016): 160-168.
- Sriraman A, Dickmanns A, Najafova Z, Johnsen SA, & Dobbelstein M. CDK4 inhibition diminishes p53 activation by MDM2 antagonists. Cell Death Dis 9 (2018): 918.
- Gao C, Xiao G, Piersigilli A, Gou J, Ogunwobi O, & Bargonetti J. Context-dependent roles of MDMX (MDM4) and MDM2 in breast cancer proliferation and circulating tumor cells. Breast Cancer Res 21 (2019): 5.
- Park SY, Jun JA, Jeong KJ, Heo HJ, Sohn JS, Lee HY, . . . Kang J. Histone deacetylases 1, 6 and 8 are critical for invasion in breast cancer. Oncol Rep 25 (2011): 1677-1681.
- de Oliveira SF, Ganzinelli M, Chilà R, Serino L, Maciel ME, Urban Cde A., . . . et al. Characterization of MTAP Gene Expression in Breast Cancer Patients and Cell Lines. PLoS One 11 (2016): e0145647.
- Milde-Langosch K, Bamberger AM, Rieck G, Kelp B, & Löning T. Overexpression of the p16 cell cycle inhibitor in breast cancer is associated with a more malignant phenotype. Breast Cancer Res Treat 67 (2001): 61-70.
- Romagosa C, Simonetti S, López-Vicente L, Mazo A, Lleonart ME, Castellvi J, & Ramon y Cajal S. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene 30 (2011): 2087-2097.
- Pandey K, An HJ, Kim SK, Lee SA, Kim S, Lim SM., . . . et al. Molecular mechanisms of resistance to CDK4/6 inhibitors in breast cancer: A review. Int J Cancer, 145 (2019): 1179-1188.
- Ocana A, & Pandiella A. Targeting oncogenic vulnerabilities in triple negative breast cancer: biological bases and ongoing clinical studies. Oncotarget 8 (2017): 22218-22234.
- Wang T, Jin X, Liao Y, Sun Q, Luo C, Wang G, . . . et al. Association of NF-κB and AP-1 with MMP-9 Overexpression in 2-Chloroethanol Exposed Rat Astrocytes. Cells 7 (2018).
- Xia Y, Shen S, & Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res, 2 (2014): 823-830.
- Engel BE, Cress WD, & Santiago-Cardona PG. THE RETINOBLASTOMA PROTEIN: A MASTER TUMOR SUPPRESSOR ACTS AS A LINK BETWEEN CELL CYCLE AND CELL ADHESION. Cell Health Cytoskelet 7 (2015): 1-10.


 Impact Factor: * 2.5
Impact Factor: * 2.5 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 