The Human Immunodeficiency Virus tat gene, its Pathogenesis and Treatment Implications
Article Information
Teto Georges*,1, Tagomo Sob Sandra2, Dambaya Beatrice1,4, Kamgaing Rachel1, Pieme constant Anatole3 and Alexis Ndjolo1,3
1Chantal Biya International Reference Centre (CIRCB), Yaoundé, Cameroon
2University of Yaoundé I, Faculty of Science
3University of Yaounde I, Faculty of Medicine and Biomedical Sciences (FMSB)
4University of Maroua, National Advanced School of Engeeniering/Department of Hydraulics and water Management
*Corresponding author: Teto Georges, Chantal Biya International Reference Centre (CIRCB), Yaoundé, Cameroon.
Received: 15 May 2024; Accepted: 21 May 2024; Published: 24 June 2024
Citation: Teto Georges, Tagomo Sob Sandra, Dambaya Beatrice, Kamgaing Rachel, Pieme constant Anatole and Alexis Ndjolo. The Human Immunodeficiency Virus tat gene, its Pathogenesis and Treatment Implications. Fortune Journal of Health Sciences. 7 (2024): 325-337.
View / Download Pdf Share at FacebookAbstract
The Human Immunodeficiency Virus type 1 (HIV-1) viral genome is 9.1 KB in length, containing both genes that code for structural, regulatory and accessory proteins found on viral single-stranded RNA. These different genes encode for specific proteins with specific functions. This is the case of the viral protein Tat, a regulatory protein, encoded by the tat gene (transactivator of transcription) of the HIV-1 viral genome, which is an essential protein for the replication, expression and progression of HIV-1 infection. It is one of the first proteins to be expressed immediately after infection. It can also penetrate into neighboring uninfected cells. It is both active in the cytoplasm and in the nucleus but more present in the nucleus. The tat gene has a length of 14 to 16 kda with two exons coding from 1 to 86 or 101 amino acids depending on the strain or isolate present (exon 1 and exon 2 necessary for its function in vivo). The multiple functions of the tat gene allow it to be a focal point of research for the understanding of the AIDS disease, the relationships and the impacts that this gene has and can develop with other cells in the context of the disease. Until date, it is the focus of several researches linking HIV-1 with other pathologies, as for example, HIV associated neurocognitive disorders, cancer oncogenesis, cardiovascular diseases; or phenomenon like viral latency establishment. The tat gene is also associated to pathologies that are induced or created, following HIV-1 infection worldwide. A literature review is thus important in order to know the current state of work on this gene which is of so much interest in HIV-1 research, including treatment and vaccine design.
Keywords
HIV, Tat, pathogenesis, treatment
HIV articles, Tat articles, pathogenesis articles, treatment articles
Article Details
Introduction
Human Immunodeficiency Virus type 1 (HIV-1) is a human retrovirus of the lentivirus genus responsible for slowly evolving pathologies grouped under the name acquired immunodeficiency syndrome (AIDS). The emergence of HIV/AIDS in 1981-1982[1], brought out a wide range of studies, particularly based on disease pathogenesis, treatment and vaccine design, in order to overcome this tremendous illness. Although continuous efforts had been made till date, there has been neither known treatment which could cure the disease nor a good vaccine for prevention or treament. In the same battle, we are focusing on this review on HIV-1 tat gene in order to bring out our contribution on HIV/AIDS studies for the future better management of HIV infected patients. HIV infection remains a major public health concern. Globally, in 2022, about 39 million people were living with HIV, and 1.3 million of people were newly infected; among them about 25 million of people were in sub-Saharan Africa [2]. Every week, 4000 adolescent girls and young women aged 15-24 years became infected with HIV globally in 2022; 3100 of these infections occurred in sub-Saharan Africa, where, women and girls (all ages) accounted for 63% of all new HIV infections [2]. In Cameroon particularly, the overall HIV/AIDS prevalence rate is about 2.7% [3], an apparently low prevalence which should emphasize the necessity of focusing on more studies in order to end AIDS by 2030 as prescribed by UNAIDS [4]. The HIV-1 viral genome is 9.1 KB in length, containing both genes that code for structural, regulatory and accessory proteins found on viral single-stranded RNA [6]. These different genes encode for specific proteins with specific functions. This is the case of the viral protein Tat, a regulatory protein, encoded by the tat gene (transactivator of transcription) of the HIV-1 viral genome, which is an essential protein for the replication, expression and progression of HIV-1 infection. It is one of the first proteins to be expressed immediately after infection, thanks to its size and its structures such as the protein transduction domain (PTD) and the nuclear localization signal (NLS) [7]. It can also penetrate into neighboring uninfected cells. It is both active in the cytoplasm and in the nucleus but more present in the nucleus. The tat gene has a length of 14 to 16 kda with two exons coding from 1 to 86 or 101 amino acids depending on the strain or isolate present (exon 1 and exon 2 necessary for its function in vivo) [8, 9]. The multiple functions of the tat gene allow it to be a focal point of research for the understanding of the AIDS disease, the relationships and the impacts that this gene has and can develop with other cells in the context of the disease. Until date, it is the focus of several researches linking HIV-1 with other pathologies induced or created, following HIV-1 infection worldwide. A literature review is thus important in order to know the current state of work on this gene which is of so much interest in HIV-1 research, including treatment.
1. Role of the tat gene in HIV-1 progression and its different functional domains identifying the viral subtypes
The tat gene
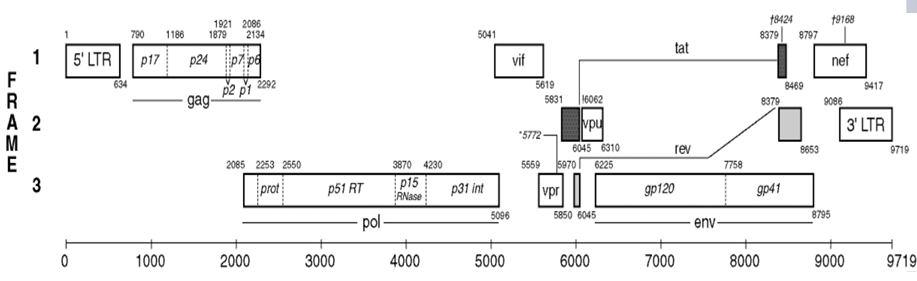
HIV-1 tat gene exists in two short different fragments of about 216 and 258 bps as colored in black in the HIV-1 gene map shown below; each of which encode a specific Tat protein. Tat-1 exon (minor form) of 72 amino acids and Tat-2 exon (major form) of 86 amino acids [11]. It acts by binding to the TAR (transactivation response) RNA element and activating transcription initiation and elongation from the LTR promoter, preventing the 5' LTR AATAAA polyadenylation signal from causing premature termination of transcription and polyadenylation. It is the first eukaryotic transcription factor known to interact with RNA rather than DNA [10].
Different functions of tat gene
The tat gene, which is part of the viral genome, has several functions during both internal and external HIV-1 infection. In addition to its main function, which is the transcriptional function allowing viral multiplication in the host organism, it also participates in HIV-1 pathogenesis. It has a regulatory, co-activating activity, it also exerts an extracellular function by affecting uninfected cells and even seeing the increased production of free radicals in the body leading to the alteration or damage of DNA thus leading to cancer.
Transcriptional function of tat gene
The tat gene performs several functions in the infected organism, but its main function remains the transactivation of the transcription of the provirus for the formation of large quantities of complete mRNAs. In the absence of the tat gene, recruitment of transcription factors to the 5' LTR promoter is sufficient to initiate transcription, but not for the production of full-length mRNAs due to premature termination of elongation and pause of RNA polymerase II (RNAPII) at the region proximal to the transcription start site [12]. The transcription of viral mRNAs resulting in the formation of new virions using the tat gene takes place in 3 steps in the nucleus of the host cell. Of which we have the initiation which is materialized by the acetylation of the nucleosomes present at the level of the 5' LTR promoter thanks to HATs (histone acetyl transferase) such as the CBP (CREB Binding Protein) / P300 complex, inducing a chromatin change allowing RNA polymerase II to transcribe viral genes [13; 14]. This rearrangement allows the recruitment of a pre-initiation complex composed of sp1, NF-κB and a TFIIH complex in order to phosphorylate the C-terminal part of RNA polymerase II (RNAPII), the polymerase is then able to transcribe from the TAR region using the multiple transcription start sites (TSS) in the promoter driven by the TATA box [15]. The RNAP II complex linked to the promoter allows the initiation of the synthesis of short mRNAs [16]. The elongation process is inhibited by two inhibitory factors such as negative elongation factors (NELF) and DRB (5,6-dichloro-1-β-D-ribofuranosylbenzimidazole) sensitivity inducing factors (DSIF). They induce the dissociation of the Pol II RNA from the proviral DNA, thus marking a pause in the elongation or stopping It [17; 12], hence the production of short mRNA strand 59 nucleotides along. The secondary structure loops of short strands of RNA formed will then bind to the TAR element (transactivation response). The formation of the TAR mRNA loop of 59 nucleotides linked to RNAP II allows the recruitment of tat at the level of the transcription complex having the capacity to remove the elongation inhibitor elements and allows the binding of Histone Acetyl transferase (HATs) such as CBP (CREB Binding Protein) / P300 at the level of the TATA box which will recruit partners for the transcription machinery (elongation). At the level of elongation, the P-TEFB complex (positive transcription elongation factor b) consisting of CDK9 and its regulatory partner cycT1 is recruited and then allows the phosphorylation of the serine residues of the C-terminal domain (CTD) of RNAPII [7]. These events allow RNAPII to achieve productive elongation and thus synthesize full-length HIV transcripts [16]. The action of tat allows the production of genomic precursors and multi and mono spliced mRNAs.
Regulatory function of the tat gene
The tat gene is a regulator of Rev-dependent mRNA transport, packaging, splicing, and translation. It also interferes with RNAi interference. The tat gene also has a stimulatory effect on reverse transcription. At high concentrations, the tat gene is able to inhibit the activity of reverse transcriptase.
Tat gene: a co-regulator of the reverse transcription
The presence of the tat gene during transcription of the provirus into mRNA, at the level of the 5'LTR promoter, involves the recruitment of transcription coactivators such as: HATs, the P-TEFb complex and chromatin remodeling [18].Thus the presence of the tat gene allows the phosphorylation of many transcription factors such as: the acetylation of the p50 subunit of NF-kb by HATs [19]. ; phosphorylation of CRP/P300 and sp120] thus allowing the activation of the regulation of the viral promoter. Sp1 is a ubiquitously expressed, prototypic C2H2-type zinc finger-containing DNA binding protein that can activate or repress transcription in response to physiologic and pathological stimuli.It has since been shown to regulate the expression of thousands of genes implicated in the control of a diverse array of cellular processes, such as cell growth. This gene itself is modified by PTMs (post translational modifications) including phosphorylation, acetylation, ubiquitination and methylation leading to the generation of the tat gene determining its binding partner for good viral transcription, it can be inhibited or activate during these PTMs [ 20, 22].
The tat gene is a regulator of the expression of certain cellular genes
It plays a role in the pathogenesis of HIV-1 by regulating in such a way as to increase the level of expression of the chemokine co-receptors CXCR4, CCR5 [23], interleukin cytokine receptors like IL-2 (CD25) in infected cells [24], IL-6, IL-10 [25]. A dysregulation of the production of cytokines contributes to the inefficiency of the immune system against the virus. This is the case of IL-10, which is a cytokine being the most anti-inflammatory, playing a far role in regulating the host's immune response to the pathogen, thus preventing damage to the host and maintaining tissue homeostasis. Normally it is produced in small quantities or even almost not in mononuclear cells. The study conducted by Bennasser et al in 2001 in human monocytes demonstrated that the Tat protein allowed an overproduction of IL-10 by activating the PKC pathway and transcription factors such as Nf-KB. This excessive production of this cytokine due to the presence of the Tat protein also leads to an increase in the calcium concentration in the monocytes by acting on the calcium signal at several levels. Moreover, Tat induces the expression of chemokines that attract uninfected T cells and macrophages, facilitating expansion of HIV in the host [26]. On the other side, Tat can also contribute to the depletion of T cells during AIDS progression by up-regulating cellular pro-apoptotic genes [27]. Finally, Tat is able to repress the transcription of several genes encoding receptors of the innate immune system responses.
HIV Integrase, a competitive element of the tat gene
Integrase (IN) is an enzyme integrating into the HIV-1 viral genome necessary for the formation of the provirus by allowing the integration of viral DNA onto DNA in the nucleus of the host cell, thus being able to be transcribed in mRNA for the formation of new virions [33]. During the transcription of the provirus, it has this capacity to be able to recruit the tat gene (transactivator of transcription) for the transcription of complete mRNAs by first binding to the TAR RNA loop. It thus has the ability to bind to both RNA and DNA [34]. Integrase (IN) being made up of the N-terminal domain, the catalytic core domain and the C-terminal domain, can, thanks to its CTD (C-terminal) domain and having a high affinity for TAR, bind to the TAR RNA loop Polymerase II. This IN-CTD TAR ARP II binding allows the recruitment of the tat gene by conformational modulation of TAR which will read to the loop. Subsequently, the gene will release IN-CTD and in turn allow the recruitment of the P-TEFB complex (CDK9 + CYCT1) which can thus phosphorylate the RNA P II at the level of the serine residues which will in turn carry out a complete transcription of the mRNAs complete [35]. It has been shown that integrase could be a competing element of the tat gene, since they have the same binding site on the TAR domain. This function that it acquires gives rise to a new therapeutic target and to the development of a new drug generation [36].
Structure of tat gene derived protein
The HIV-1 tat gene encodes a 14 to 16 kda Tat protein with two exons ranging from 1 to 86 or 101 amino acids depending on the isolates present. Exon 1 ranges from 1 to 72 amino acids essential to its in vivo and in vitro function, including the transactivation of the highly conserved viral promoter and exon 2 from 73 to 86 or 101 amino acids essential to its function in vivo, poorly conserved [37; 38]. Based on previous studies, researchers have been able to demonstrate that the Tat protein has five major functional domains. The first four are located in the first exon (I, II, III, IV) all having very specific functions and of domain V in the second exon. Domain I is involved in protein structuring (1 to 21 aa), cysteine-rich domain II is required for tat transactivation, domain III or 'core' is highly conserved and is essential for the transactivator function of Tat, a mutation at the level of the latter leads to the abolition of transcriptional activity. Domain IV is the basic domain of the protein. It allows attachment to the TAR sequence of the newly synthesized viral RNA and also contains the nuclear localization signal (NLS, aa 48 to 59) which contributes to localization of Tat in the nucleus where it exerts its transactivating function [33].
Variations of the functional domains of tat gene according to viral subtype
The Tat protein encoded by the tat gene is involved in the progression of HIV-1 in the host cell, due to its multiple functions within the virus and in infected cells. The Tat protein plays several roles such as replication of the provirus (necessary for the formation of new virions), infection of neighboring cells, involvement in the immune response, it is involved in the regulation of certain genes, in the affection of certain specific cells such as neurons, playing a role in the production of free radicals that can lead to the development of opportunistic diseases such as cancers. These different functions clearly demonstrated from the different scientific studies show clearly the implication of the tat gene in the progression of the HIV-1 disease leading to AIDS. In the serum of infected patients, there are viral RNA and proteins among which the Tat protein. Since the level of Tat antibodies found in it is high, research has led to a thought of setting up a prophylactic vaccine against this Tat protein [28]. Animal and preclinical studies on subtype B have shown efficacy in inducing a Th1 and Th2 immune response, thereby increasing immune function in these patients. The Human Immunodeficiency Virus exists in two types, type 1 and type 2. HIV-1 is the most prevalent form in the world, accounting for 95% of all HIV-related infections [29]. HIV-1 exists in the world in four major groups: group M (the main one), group O (the outliers), group N (the new ones) which are non-M and non-O and the rarest group P. Due to its spread and phylogeny, group M is subdivided into 9 subtypes of A-D, F-H, J and K and unique recombinant forms [30], one hundred and fiftyseven circulating recombinant forms (CRF157_A6C) and several unique unclassified recombinant forms (URFs) have been identified to date. The subtypes are in turn subdivided into other subtypes of which for the most part we have A1, A2 and F1, F2. There are also subtypes that recombine with each other, for example CRF01-AE, a recombinant form of the A and E subtypes (probably ancestral and not yet described) of group M. [31]. This high genetic variability is due to a high rate of mutations and a high rate of inter- and intra-molecular recombination that occurs in infected host cells due to a lack of DNA correction or proofreading system during reverse transcriptase action [32]. From the study of the genetic disposition of HIV-1 in the world, we note that 50% of the infections are caused by the C subtype generally found in Sub-Saharan Africa, India and South America. The most studied clade, subtype B, accounts for 10% of infections and is most commonly found in Europe and America. Subtypes A and D are found in Sub-Saharan Africa with a percentage of 12% and 3% respectively and subtype G of 6%; subtypes F, H, J and K represent 0.94% of infections. The recombinant circulating forms CRF01_AE based in East Asia and CRF_AG represent 5% of infections worldwide and CRF03_AB represents 0.1% [30]. It is also stated that the other forms represent the remaining 8% of infections. All recombinant forms combined are responsible for 18% of infections worldwide [30]. Beside these HIV-1 subtypes, CRFs and URFs, there is the first CRF namaly CRF01_AB spreading in Japan [5].
Study of the variability of the tat gene using genomic sequencing shows a variation of the sequences of the tat gene belonging to different subtypes. The study conducted by Teto et al in 2016 on 100 Cameroonian HIV positive samples showed genetic variability of the tat gene of different subtypes. From this study, several recombinant forms were detected in the study area and different mutations in the different existing subtypes were: N29K and N36I on CRF02_AG and CRF11_cpx isolates respectively; N23T, N24K, and K29T subtype G isolate; N23T and T24K on CRF13_cpx; K24S, Y26H, and K29H on subtype D; N24K and F26W on CRF22_01A1; K24Q, C31S, and P35Q on CRF18_cpx; and N23S and K29R on the CRF01_AE isolates [29]. No mutations were observed in the cysteine region of CRF37_cpx. In 93% of the samples analyzed, only three CRF02_AG isolates showed mutations at C31 (C31S/A/F) and one CRF18_cpx isolate showed a C31S mutation and one CRF18_cpx isolate showed a C31S mutation. Several CRF02_AG samples showed mutations from asparagine (N) to lysine (K) in the cysteine-rich and Core regions, including N24K, N29K, and N40K in 44%, 58%, and 30% of samples, respectively. All subtype G samples also showed an N24K mutation and 42% of CRF02_AG samples had an S23N substitution. Compared to the consensus sequences, no major mutations were observed in the N-terminal Tat region of Cameroon isolates except for the P3L substitution in 63% of CRF02_AG isolates. These mutations can lead to functional modifications of the different domains of the Tat gene. Lysine-associated hydrogen bonds are important for protein stability and K residues play an important role in HIV-1 Tat transactivation. Studies of HIV-1 B, C and CRF01_AE Tat subtypes have shown that, compared to B Tat subtype, there was a significant increase in viral transactivation with CRF01_AE and C Tat subtype [27]; K residues in the cysteine-rich, Core, and TAR-binding regions played an important role in this increased Tat activity and viral transactivation, and mutations in K residues in A decreased viral transactivation by two to 20 folds [27]. Thus, it is possible that mutations resulting in increased K residues, as shown in the Cameroon study for CRF02_AG and G subtype isolates, could result in increased LTR transactivation and replication in individuals infected with these subtypes. This said, a large variability in the functional domains of the gene tat viral subtypes implies a modification of the interaction of the viral transactivator with cellular and/or viral proteins influencing the overall level of transcriptional activation as well as its action as a neurotoxic protein. Therefore, genetic variability within Tat may impact the molecular architecture of Tat protein functional domains that may impact HIV pathogenesis and disease [28].
Tat gene and HIV-associated neurocognitive disorders (HAND)
HIV is the target of brain mononuclear phagocytosis (monocyte-derived macrophage and microglia) and immune suppression accelerates virus growth and associated neuronal damage. One of the pathologies related to HIV is the condition of the central nervous system which affects cognitive, behavioral and motor function which triggers a range of negative clinical outcome. There are nearly 27% of people infected with HIV-1 who have identifiable cognitive dysfunction and 84% who have definable deficits in cognition, physiology and behavior [40]. The tat gene (transactivator of transcription), which is involved in the transcription machinery of the HIV provirus type 1 in the host cell, is also a powerful mediator involved in the development of neurocognitive disorders associated with HIV-1 (HAND). Despite taking antiretrovirals, it is able to cross the blood-brain barrier and finally attack the central nervous system (CNS). It interacts at this level with microglia, astrocytes, microvascular endothelial cells of the brain and neurons [41].
HIV-1 by itself cannot infect neurons in general, but lesions and excitotoxicities on CNS neurons suggest HIV-1 infection of other CNS cells such as microgolia, atrocytes [42] which can produce and secrete viral proteins such as the Tat protein and pro-inflammatory cytokines which can attack neurons even in the absence of active HIV-1 replication [43]. HIV-1 has four major groups, of which the M form is the most widespread and includes subtypes ranging from A-D, F-H, J and K [44]. It is noted that subtype B is the most predominant. Studies have shown a genetic, biological and pathological difference in the tat gene of subtype B and subtype C, that of subtype B being 101 amino acids in length. The extracellular tat gene has this ability to infect other cells such as the CNS, this recombinant and purified form acts according to the subtype belonging to the length of the gene. Thus, from reviews of articles studying the impact of the tat gene belonging to different subtypes and having different lengths on the central nervous system in laboratory rats, it emerges that the most found and studied protein of 101 Amino helpers including Tat variants, such as Tat 72 and Tat 86 exhibit a number of distinct activities with respect to mediating CNS damage and neurotoxicity. The following figure presents the different functionalities of variants of the Tat protein in the induction of neurocognitive diseases associated with HIV-1.
The Tat protein, which is a mediator of neurocognitive diseases, acts on cells of the central nervous system as well as neuron receptors, thus playing several roles that contribute to the production of neurotoxicity in patients carrying the HIV-1 virus. The Tat protein released by these immune cells from central nervous system phagocytosis called extracellular tat, combined with free radicals (which can be produced by the tat gene) attacks neurons by binding to neuron receptors such as NMDAR (N-Methyl -D-aspartate), LRP or CXRC4 allowing endocytosis. This leads to excitotoxicity and apoptosis by increasing by potentiation of glutamate through the NMDAR pathway, hence the destruction of neurons [42].
Involvement of the tat gene in cancer oncogenesis
Thanks to its size (14kDa) and its structures (the protein transduction domain and its cell localization signal), the tat gene is able to penetrate uninfected neighboring cells and can create metabolic disturbances at this level such as increased production of oxygenated species and the production of oncogenesis leading to cancers. The incidence of neoplasms increases drastically in HIV-1 infected patients, even when on long-term treatment. Several types of cancer have been associated with HIV-1 infection and AIDS, such as Kaposi sarcoma (KS), lymphoproliferative disorders and cervical cancer [45]. Cervical cancer is the most common cancer in developing countries and affects most women with HIV-1, and is recurrent in women with HIV-1 and not dependent on immunosuppression [46]. Kaposi's sarcoma one of the malignant cancers occurring in HIV-1 patients is the most common tumor, it usually occurs in the late stage of severe immunosuppression [47]. It is characterized by the proliferation of spindle cells that appear to be of lymphatic endothelial origin. The extracellular Tat protein can infect neighboring cells and is able to enter endothelial cells, which are one of its targets. Tat protein induces angiogenesis in endothelial cells and the growth of KS spindle cells [48; 49]. Overexpression of Tat in vivo leads to an increase in KS-like lesions, so Tat acts as a cofactor in KS production [50] although its mechanism of action is not yet well elucidated. B-cell lymphoma is another type of cancer occurring in HIV-1 patients. The tat gene induces at this level an increased production of ROS leading to oxidative DNA damage which is a source of genomic instability thus activating the PI3K /AKT/ mTORC1 (phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin complex 1) pathway which in turn inhibits the expression of cMYB and E2F8 (E2F transcription factor 8), being transcriptional inhibitors of AICDA (activation-induced cytidine deaminase). This inhibition promotes overexpression of this gene, resulting in the oncogenic translocation between MYC (The oncoprotein MYC is a pleiotropic transcription factor that modulates global gene expression and regulates critical cellular processes including proliferation, differentiation, cell cycle, metabolism and apoptosis.) and IGH characteristic of Burkitt's lymphoma [51]. Note that the tat gene allows the reconciliation of the MYC and IGH loci in the B lymphocyte initially located in different nuclear compartments. The Tat protein encoded by the tat gene is a true oncogenic cofactor in patients carrying this virus.
The role of HIV-1 Tat in HIV-1 related cardiovascular disease
Several studies have shown that most HIV-1 patients also suffer from cardiovascular disease. This is due to factors such as dyslipidemia, chronic inflammation, abnormal lipids such as HDL, increased triglycerides and the duration of antiretroviral therapy taken by HIV-positive patients [52, 53, 54]. HIV-associated cardiovascular disease is characterized by cardiomiopathy, increased fibrous tissue and significant infiltration of immune cells in the heart [52]. Several studies have demonstrated that gp120 and Tat proteins are involved in the development of cardiovascular disease in HIV patients. Through certain pathways, these proteins lead to apoptosis of cardiomyocytes and epithelial cells. The MAPK/ERK Kinase pathway (MEK inhibition) (mitogen-activated protein kinase/ ERK ist the first mammalian MAP kinase) inhibits apoptosis in cardiac cells [55, 56]. The role of Tat protein in HIV-associated cardiovascular disease is increasingly recognized. Studies have shown that Tat protein induces transcriptional changes in different cell types that make up the heart, and some of these transcriptional changes are similar to those observed in certain non-HIV-associated cardiovascular diseases [57]. Tat protein induces connexin 43 (Cx43) mRNA and protein in cardiomyocytes and increased levels of lipofuscin, a known biomarker of cardiac aging. Both of these observations are also seen in non-HIV-related cardiovascular disease [52]. Interestingly, in human hearts, an increase in Cx43 protein expressed in response to Tat exposure has also been observed, suggesting that Cx43 undergoing Tat exposure is more sensitive to the effects of Tat exposure [58]. Tat-induced cardiomyocyte apoptosis led to the formation of fibrous tissue plaques and the accumulation of mitochondria. This suggests that cardiomyocyte apoptosis is a mitochondria-controlled pathway. High levels of circulating ATP correlate with cognitive impairment and endothelial damage [59]. Furthermore, exposure of cardiomyocytes to an increasing concentration of ATP in vitro caused an increase in the frequency of pain and inflammation. These results show that HIV infection and exposure to viral proteins result in damage to the heart and cardiovascular disease.
Tat protein and inhibition of latency establishment
Most of the world's HIV patients are treated with highly active antiretroviral drugs to increase CD4+ T cells and reduce viral load. Infected CD4+ T cells lie dormant in the body, preventing complete eradication of the disease in these patients [60]. The latent reservoir is established during acute infection and represents an archive of wild-type and drug-resistant viruses [61]. Reactivation of latently infected cells is probably the main source of viral rebound when treatment is interrupted or terminated [62]. Thus, HIV-1 latency allows infection to persist throughout life, and is the subject of intense research. Several approaches aimed at reactivating the latent reservoir have been used in clinical trials. The establishment of HIV-1 latency is mainly the result of one or more transcriptional blockages, usually at the time of infection of an activated CD4+ T cell that switches to the resting state. NF-B (Nuclear Factor K-B) and/or Nuclear Factor of Activated T Cell (NFAT) (depending on the cell type) are required to initiate viral transcription by binding to the activated CD4+ T cell. The Tat protein, important for viral transcription and multiplication in the host, also plays a key role in viral latency. There are several mechanisms by which Tat protein, if present in sufficient quantities, could counteract the establishment of HIV-1 latency by promoting transcription initiation or elongation. The transcriptionally active form of NF-B, p50/p65, can be sequestered in the cytoplasm by IκBα. Tat itself can induce nuclear translocation of NF-B p50/p65 [63], probably via a direct interaction with Protein Kinase Reductase (PKR) (double-stranded). This may lead to IκBα degradation [64]. IκBα(NuclearFactor ofKappa light polypeptide gene enhancer inB-cells Inhibitoralpha;NFKBIA) is one member of a family of cellular proteins that function to inhibit theNF-κBtranscription factor. IκBα inhibits NF-κB by masking thenuclear localization signals(NLS) of NF-κB proteins and keeping them sequestered in an inactive state in the cytoplasm.In addition, IκBα blocks the ability of NF-κB transcription factors to bind to DNA, which is required for NF-κB's proper functioning. In addition to initiating transcription, NF-B p50/p65 can displace HDAC1 (deaceted histone). The p50/p50 homodimers bound to HDAC1 (histone deacetylase 1) at the B sites of the viral promoter. Chromatin is also regulated by the Tat protein through restrictive modifications. Histone acetyltransferases (HATs), notably p300, CBP and PCAF, are recruited by Tat to the 5' LTR [65], where they can reverse the effects of histone deacetylation. Nucleosome remodeling is induced by Tat via recruitment of the Ini1, BRG-1 (Brahma-related gene-1) and Brm components of the SWI/SNF chromatin remodeling complex (The mammalian SWI/SNF complexes mediate ATP-dependent chromatin remodeling processes that are critical for differentiation and proliferation.) [14, 66] and by recruitment of the histone chaperone hNAP-1 (human Nucleosome Assembly Protein-1) [67], which relaxes chromatin structure and thus enables transcription. Tat can also overcome elongation barriers by disrupting the RNP 7SK (human 7SK ribonucleoprotein) complex through direct displacement of Hexim 1 resulting in increased nuclear levels of enzymatically active P-TEFb [68, 69]. Hexim 1 is an essential transcription regulator during Human erythropoiesis. Hexamethylene bis-acetamide inducible 1 (HEXIM1) regulates RNAPII activity by controlling the location and activity of positive transcription factor β. Findings show that in vivo depletion of the tat gene contributes to the establishment of virus latency. One example is the study carried out by Daniel A. and colleagues in 2012, where the in vitro menes study on cells and virus clearly shows that the presence of large quantities of intracellular Tat leads to inhibition of virus latency. Good therapeutic follow-up is important in the establishment of latency, thus preventing the presence in sufficient quantity of the Tat protein which plays a primordial role in the activation of latency [70].
Tat as a therapeutic target
A number of drugs have been developed for the management of patients with HIV worldwide, with the aim of reducing viral load and increasing TCD4+ cell counts. These drugs are known as antiretroviral drugs, and include among others, protease inhibitors, nucleoside reverse transcriptase inhibitors, non-nucleoside transcriptase inhibitors and integrase inhibitors. With proper therapeutic follow-up, these ARVs allow HIV latency in seropositive patients; if treatment is interrupted, patients will switch from the resting state of the virus to the active state with the help of the tat gene [71, 72]. Recent studies have shown that these ARVs cannot inhibit the transcription of the virus that is driven by the HIV-1 tat gene. This tat gene has been the subject of several studies and has been clearly shown to be involved in several mechanisms leading to disease progression and commorbidity in patients with the virus [73, 74]. Given its involvement in the progression of this incurable disease, researchers have focused on this protein as a potential therapeutic target for improving patient care. As a result, anti-Tat agents have been developed for the treatment of HIV. The review by Hongping et al in 2020 clearly shows that through certain mechanisms including the Tat protein, it is possible to establish a vaccine using anti-Tat. They suggest that the development of an agent that inhibits newly synthesized Tat and thus virus production by infected cells would be an important step towards a functional cure [75,76]. Tests with Nullbasic and HT1 have shown that both can strongly inhibit HIV transcription, and the former has been tested in mouse models of acute HIV infection. Their application would most likely require a gene therapy approach in hematopoietic stem cells. The question of whether immune cells protected against HIV infection by Nullbasic or HT1 would escape pre-existing humoral and cellular immune responses to Tat. However, a Phase I/II clinical trial of a Rev TDN (Trans-Dominant Negative) called RevM10 showed stable expression for over 100 weeks and a preference for Tat 168. TDN inhibitors (TDNi) are defined as a mutant protein that lack an intrinsic activity that can inhibit function of the wild-type protein intrans. Early studies showed that Tat TDNi inhibited HIV-1trans-activation of transcription if expressed in excess compared to wild-type Tat. Didehydro-cortistatin A (dCA) has the remarkable ability to inactivate Tat and preclinical studies suggest that it can "lock" the HIV promoter so that RNAP II-mediated transcription is durably inhibited, even after dCA treatment is stopped. However, large-scale production of dCA may prove difficult [77], which could limit its availability. As the molecular interaction between dCA and Tat has been modeled [78], future in silico screening experience using pharmacopoeia-based models of the interaction of dCA with Tat could identify new agents with anti-Tat activity. Triptolide, a drug used to treat rheumatoid arthritis, has been converted into an HIV inhibitor and is awaiting the results of clinical trials in terms of safety and efficacy. Triptolide affects global transcription, which may limit its usefulness in the fight against HIV.
Tat protein as vaccine target
Vaccination has been successful in control of various viral diseases and induces specific cytotoxic T-cell elimination of infected cells displaying viral proteins in association with HLA molecules and/or specific antibody blocking and clearing of free virus. This approach is effective for viruses with stable phenotypes, such as smallpox and measles, and for viruses with limited variation in their antigenic epitopes, such as poliomyelitis. But this mode of vaccination becomes more problematic with viruses such as influenza, for which the predominant epitopes may change from year to year, necessitating the preparation of an annual vaccine for use before the winter flu season. For HIV-1, the huge diversity in immunogenic viral epitopes and the rapid mutational variations that occur within and between individuals' have so far prevented successful application of these conventional approaches [79], so far, HIV Tat can offer an alternative to this; first, the two tat immunodominant domains in the essential first exon of Tat (amino acids 1-72) are relatively conserved, no doubt because of restraints imposed by overlapping reading frames. Antibodies to each of two separate immunogenic domains can inhibit cellular uptake, and the polyclonal response in a Tat-vaccinated subject should yield a variety of antibody binding patterns with a high probability of uptakeblocking antibodies to either or both of the immunodominant domains in the majority of extracellular Tat proteins. Second, specific antibody interdiction of Tat in the extracellular fluid would inhibit the replication of all HIV-1 quasispecies indiscriminantly and should thus exert no pressure for the selection of variants producing particular Tat proteins. Therefore, selective development of quasispecies producing antibody unreactive to Tat proteins should not occur within a given individual. Third, prevention of massive virus replication must minimize opportunities for the development of mutant viruses with variant structural proteins permissive for evasion of the earlier antiviral immune response. A further corollary of this analysis is that, to be successful, antibody interdiction of the predominant populations rather than all variants of Tat proteins should suffice to minimize the initial rapid burst of viral expansion. it is envisaged that such a Tat vaccine would be used alone, in a manner analogous to toxoid vaccination for tetanus and diphtheria [80, 81]. Antibodies to Tat occur both naturally and in response to HIV-1 infection, and there is a correlation of low or absent antibodies with progression to AIDS [82-83], further supporting the notion that extracellular Tat interdiction by antibodies can contribute to the control of HIV-1 infection. Tat vaccination represents an example of a “pathogenic-driven” intervention that is potentially effective for both preventative and therapeutic strategies, since it is aimed at blocking virus transmission and spread. The rationale is based on the evidence that HIV-1 Tat, which is necessary for HIV gene expression, replication, and cell-to cell transmission, appears also to be critical in the initial steps of virus acquisition.
Ensoli and al showed that a therapeutIc Tat vaccine was able to promote increases of CD4+ T-cells and return to immune homeostasis while reducing the virus reservoir in chronically cART-treated patients in a similar way to that seen in spontaneous 'post-treatment controllers' whose immune system can control the reactivation of HIV after discontinuing therapy. They showed that such kind of vaccine was an optimal vaccine candidate for cART intensification towards HIV reservoirs depletion, functional cure, and eradication strategies [84-85].
Conclusion
The Tat protein, encoded by the Human Immunodeficiency Virus tat gene, is a gene with multiple functions in HIV disease, creating co-morbidity in patients. Its main role is in the transcription of proviral DNA leading to viral multiplication, but it is also involved in several mechanisms leading to severe disease and activation of virus latency. Cases of commorbidity include its involvement in cardiovascular disease, cancer and neurocognitive disorders in HIV-positive patients. Tat protein is a potential therapeutic target for improving the health of these patients. Particular interest is focused on this gene, given its importance and role in several mechanisms affecting HIV-positive patients. With a view to improving patient care, researchers should focus more on inhibiting this gene to counteract its harmful effects in these patients, althought most efficient conclusions has not yet been drive in term of therapy or vaccine, but hopefully, by focusing all our attention on it, maybe a better solution could be found on tat gene studies in a shortcoming future.
Conflict of interest
Authors declare no conflict of interest
Funding
This work was supported by the Chantal Biya International Reference Centre for HIV/AIDS research and prevention.
References
- Amat-Rose JM. L’infection à VIH et le SIDA en Afrique noire: facteurs d’épidémisation et de régionalisation. Portail des revues scientifiques en SHS 42 (1989): 333- 355.
- UNAIDS 2023. Global HIV statistics, factsheets (2022): 1-6.
- CNLS 2022. Profil des estimations et projection en matiere de VIH/SIDA au Cameroun, CNLS (2022): 9-12.
- UNAIDS 2023. The path that end AIDS (2023): 1-12.
- Shiro Ibe,Yoshiyuki Yokomaku,Teiichiro Shiino,Rie Tanaka,Junko Hattori,Seiichiro Fujisaki,et al. HIV-2 CRF01_AB: first circulating recombinant form of HIV-2. J Acquir Immune Defic Syndr 54 (2010): 241-7
- Stoltzfus CM and Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res 4 (2006): 43-55.
- Ne E, Palstra RJ, Mahmoudi T. Transcription: Insights From the HIV-1 Promoter. Int. Rev. Cell Mol. Biol 335 (2018): 191-243.
- Neuveut C andJeang KT. Recombinant human immunodeficiency virus type 1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J Virol 70 (1996): 5572-81.
- Peloponese JM Jr, Gregoire C, Opi S, Esquieu D, Sturgis J, Lebrun E, et al. 1H-13C nuclear magnetic resonance assignment and structural characterization of HIV-1 Tat protein. C R AcadSci III 323 (2000): 883-94.
- https://www.hiv.lanl.gov/content/sequence/HIV/COMPENDIUM/2000/partI/Karn.pdf. Tat, a novel regulator of HIV transcription and latency, reviews. Accessed 03-10-2023.
- https://www.hiv.lanl.gov/content/sequence/HIV/MAP/landmark.html. HIV-1 gene map. Accessed 03-10-2023.
- Natarajan, M. and al. Negative elongation factor (NELF) coordinates RNA polymerase II pausing premature termination, and chromatin remodeling to regulate HIV transcription. J. Biol. Chem 228 (2013): 25995-260003.
- Deng, L., de la Fuente, C., Fu, P., Wang, L., Donnelly, R., Wade, J. D., Lambert, P., Li, H., Lee, C. G. and Kashanchi, F. Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology 277 (2000): 278-95.
- Pumfery, A., Deng, L., Maddukuri, A., de la Fuente, C., Li, H., Wade, J. D., Lambert, P., Kumar, A. and Kashanchi, F. Chromatin remodeling and modification during HIV-1 Tatactivated transcription. Curr HIV Res 1 (2003): 343-62.
- Kadonaga, J. T. Perspectives on the RNA polymerase II core promoter. Wiiley internatonal discip. Rev. Dev. Biol 1 (2012): 40-51.
- Williams, S. A., Chen, L. F., Kwon, H., Ruiz-Jarabo, C. M., Verdin, E. and Greene, W. C. NFkappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. Embo J 25 (2006): 139-49.
- Zhang, Z. and al. Negative elongation factors NFEL represses human immunodeficiency virus transcription by pausing the RNA polymerase II complex. J. Biol. Chem 282 (2007): 16981-16988.
- Ott M, Geyer M, Zhou Q. The Control of HIV Transcription: Keeping RNA Polymerase II on Track. chom 11 (2011): 2-12
- Furia B, Deng L, Wu K, Baylor S, Kehn K, Li H, Donnelly R, Coleman T, Kashanchi F. Enhancement of Nuclear Factor-kB Acetylation by Coactivator p300 and HIV-1 Tat Proteins. Issue of February 277 (2002): 4973-4980.
- Rossi A, Mukerjee R, Ferrante P, Khalili K, Amini S, Sawaya B E. Human immunodeficiency virus type 1 Tat prevents dephosphorylation of Sp1 by TCF-4 in astrocytes. Laboratory of Biology, Don C. Gnocchi Foundation, IRCCS, 20148 (2006): 12-25
- Zhang, H. S., Chen, X. Y., Wu, T. C., Sang, W. W., and Ruan, Z. MiR-34a is involved in Tat-induced HIV-1 long terminal repeat (LTR) transactivation through the SIRT1/NFkB pathway. FEBS Lett 586 (2012): 4203-4207.
- D’Orso I, Frankel A, D. HIV-1 Tat: Its Dependence on Host Factors is Crystal Clear. Department of Biochemistry and Biophysics, University of California 600 (2010): 94158-2280.
- Huang, L., Bosch, I., Hofmann, W., Sodroski, J., and Pardee, A. B. Tat protein induces human immunodeficiency virus type 1 (HIV-1) coreceptors and promotes infection with both macrophage-tropic and T-lymphotropic HIV-1 strains. J. Virol 7 (1998): 8952-8960.
- Rayne, F., Debaisieux, S., Yezid, H., Lin, Y. L., Mettling, C., Konate, K., and al. Phosphatidylinositol-(4,5)-bisphosphate enables efficient secretion of HIV-1Tat by infected T-cells. Embo J 29 (2010):1348-1362.
- Bennasser Y., Contreras X., Moreau M., Leclerc C., Badou A. and Elmostafa B. La protéine Tat du VIH-1 induit la production d’IL-10 par le monocyte humain : implication de la voie PKC et de la voie calcique. Journal de la Société de Biologie. 195 (2001): 319-326
- Elena S Izmailova, Frederic M.N. Bertley, Norbert Makori, Qian Huang. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nature Medicine 9 (2003): 191-7.
- Kim N, Kukkonen S, Gupta S, Aldovini A, Rice AP. Association of tat with promoters of PTEN and PP2A subunits is key to transcriptional activation of apoptotic pathways in HIV-infected CD4+ t cells.PLoS Pathogens 6 (2010): e12686.
- Goldstein G, Damiano E, Donikyan M, Pasha M, Beckwith E and Chicca J. HIV-1 Tat B-cell epitope vaccination was ineffectual in preventing viral rebound after ART cessation. HIV rebound with current ART appears to be due to infection with new endogenous founder virus and not to resurgence of pre-existing Tat-dependent viremia. Hum Vaccin Immunother 10 (2012): 1425-12
- Teto G, Fonsah J Y, Tagny C, Mbanya D, Nchindap E, Kenmogne L, Fokam J, Njamnshi D, Kouanfack C, Njamnshi A, and Kanmogne G. Molecular and Genetic Characterization of HIV-1 Tat Exon-1 Gene from Cameroon Shows Conserved Tat HLA-Binding Epitopes: Functional Implications Viruses 8 (2016): 1-8
- Desfosses, Y.; Solis, M.; Sun, Q.; Grandvaux, N.; van Lint, C.; Burny, A.; Gatignol, A.; Wainberg, M.A.; Lin, R.; Hiscott, J. Regulation of human immunodeficiency virus type 1 gene expression by clade-specific tat proteins. J. Viro 179 (2005): 9180-9191.
- Li L, Dahiya S, Kortagere S, Aiamkitsumrit B, Cunningham D, Pirrone V, Nonnemacher M and Wigdahl B Impact of Tat Genetic Variation on HIV-1 Disease. Adv virol 10 (2012): 1-28
- Samri, A., Charpentier, C. and Cheynier, R. Le réservoir viral dans l’infection par le VIH-2, modèle d’une infection rétrovirale atténuée. Medecine science 36 (2020): 336-339.
- Cecilia Rocchi, Camille Louvat, Adriana Erica Miele, Julien Batisse, Christophe Guillon, Lionel Ballut, Daniela Lener, Matteo Negroni, Marc Ruff, Patrice Gouet and Francesca Fiorini. The HIV-1 Integrase C-Terminal Domain Induces TAR RNA Structural Changes Promoting Tat Binding. International Journal of Molecular Sciences.Int. J. Mol. Sci 23 (2022): 3742.
- Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 20 (2006): 13-23
- Eberle, J., and Gutler, L. Basic principles of HIV HIV Types, Groups, Subtypes and Recombinant Forms: Errors in Replication, Selection Pressure and Quasispecies.10 (2012):79-83.
- Palaniappan, C., Wisniewski, M., Wu, W., Fay, P.J., Bambara, R.A. Misincorporation by HIV-1 reverse transcriptase promotes recombination via strand transfer synthesis. J. Biol. 271 (1996): 22331-22338.
- Maertens, G.N.; Engelman, A.N.; Cherepanov, P. Structure and function of retroviral integrase. Nat. Rev. Microbiol. 20 (2022): 22-34.
- Jeang, K. T., Xiao, H. and Rich, E. A. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. J Biol Chem 274 (1999): 28837-40.
- Evan Clark, Brenda Nava, and Massimo Caputi. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget 16 (2017): 27569-27581.
- Hetzer, C., Dormeyer, W., Schnolzer, M. and Ott, M. Decoding Tat: the biology of HIV Tat posttranslational modifications. Microbes Infect 7 (2005): 1364-9
- Debaisieux, S., Rayne, F., Yezid, H. and Beaumelle, B. The ins and outs of HIV-1 Tat. Traffic 13 (2012): 355-63
- El-Amine R, Germini D, Zakharova V V., Tsfasman T, Sheval E V., Louzada RAN, Dupuy C, Bilhou-Nabera C, Hamade A, Najjar F, and al. HIV-1 Tat protein induces DNA damage in human peripheral blood B-lymphocytes via mitochondrial ROS production. Redox Biol 15 (2018): 97-108.
- Heaton, R.K. and al. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J. Neurovirol 17 (2011): 3-16.
- Marino, J., Maubert, M.,Mele, A., Spector, C., Wigdahl, B. and Nonnemacher, M., Functional impact ofHIV-1 Tat oncells oftheCNS andits role inHAND. Cellular and Molecular Life Sciences. 24 (2020): 5079-5099.
- Nookala AR., Kumar A. Molecular mechanisms involved in HIV-1 Tat-mediated induction of IL-6 and IL-8 in astrocytes. J Neuroinflammation. 11 (2014): 214.
- Tatro ET, Soontornniyomkij B, Letendre SL, Achim CL. Cytokine secretion from brain macrophages infected with human immunodeficiency virus invitro and treated with raltegravir. BMC Infect Dis 236 (2014): 1471-2334.
- Liner KJ 2nd, Hall CD, Robertson KR. Impact of human immunodeficiency virus (HIV) subtypes on HIV-associated neurological disease. J Neurovirol. 13 (2007): 291-304.
- Aoki Y, Tosato G. Targeted inhibition of angiogenic factors in AIDS-related disorders. Current Drug Targets - Infectious Disorders 3 (2003): 115-128.
- Clarke B, Chetty R. Postmodern cancer: the role of human immunodeficiency virus in uterine cervical cancer. Molecular Pathology 55 (2002): 19-24.
- Nasti G, Tirelli U. Highly active antiretroviral therapy in AIDS associated Kaposi's sarcoma (KS): implications for the design of therapeutic trials in patients with advanced symptomatic KS. Journal of Clinical Oncology 23 (2005): 2433-2434
- Aoki Y, Tosato G. Neoplastic conditions in the context of HIV1 infection. Current HIV Reseach 2 (2004):343-349
- Rusnati M. Presta M. HIV-1 Tat protein and endothelium: from protein/cell interaction to AIDS-associated pathologies. Angiogenesis 5 (2002): 141-151.
- Weiss R, Boshoff C. Addressing controversies over Kaposi's sarcoma. Journal of the National Cancer Institute 92 (2000): 677-679.
- Burkitkan A, Germini D, Bissenbaev A. K, Musinova Y. R, Scheval E. V, Vassetzky Y and Dokudovskaya S. HIV-1 Tat Activates Akt/mTORC1 Pathway and AICDA Expression by Downregulating its Transcriptional Inhibitors in B Cells. J. Mol. Sci.22 (2021): 1588.
- Sun Y, Huang YC, Xu QZ, Wang HP, Bai B, Sui JL, Zhou PK. HIV-1 Tat depresses DNA-PK(CS) expression and DNA repair, and sensitizes cells to ionizing radiation. International Journal of Radiation Oncology, Biology, Physics 65 (2006): 842-85
- Prevedel, L., Morocho, C. M., Bennett, V. L., and Eugenin, E. A. HIV associated cardiovascular disease: role of connexin 43. Am. J. Pathol 187 (2017): 1960-1970.
- Palma Reis, R. Cardiovascular risk in HIV-infected patients. Port. Cardiol 38 (2019): 471-472.
- Estrada, V., Domingo, P., Suarez-Lozano, I., Gutierrez, F., Knobel, H., Palacios, R., and al. Risk of cardiovascular disease in patients with HIV infection undergoing antiretroviral therapy. Rev. Clin 220 (2020): 149-154.
- Lee, E. O., Kim, S. E., Park, H. K., Kang, J. L., and Chong, Y. H. Extracellular HIV-1 Tat upregulates TNF-α dependent MCP-1/CCL2 production via activation of ERK1/2 pathway in rat hippocampal slice cultures: inhibition by resveratrol, a polyphenolic phytostilbene. Exp. Neurol 229 (2011): 399-408.
- Cipolletta, E., Rusciano, M. R., Maione, A. S., Santulli, G., Sorriento, D., Del Giudice, C. and al. Targeting the CaMKII/ERK interaction in the heart prevents cardiac hypertrophy. PLoS One 10 (2015):
- Zhao, G., Qiu, Y., Zhang, H. M., and Yang, D. Intercalated discs: cellular adhesion and signaling in heart health and diseases. Heart Fail Rev 24 (2019): 115-132.
- Schultz, F., Swiatlowska, P., Alvarez-Laviada, A., Sanchez-Alonso, J. L., Song, Q. A., and Gorelik, J. Cardiomyocyte-myofibroblast contact dynamism is modulated by connexin-43. Faseb J 33 (2019): 10453-10468.
- Seror, C., Melki, M. T., Subra, F., Raza, S. Q., Bras, M., Saidi, H., and al. Extracellular ATP acts on P2Y2 purinergic receptors to facilitate HIV-1 infection. J. Exp. Med. 208 (2011): 1823-1834.
- Pomerantz RJ. Reservoirs of human immunodeficiency virus type 1: the main obstacles to viral eradication. Clin. Infect. Dis 34 (2002):91-97.
- Hen L, Siliciano RF. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection. J. Allergy Clin. Immunol 122 (2008): 22-28.
- Joos B, and al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc. Natl. Acad. Sci 105 (2008): 16725-16730
- Demarchi F, d’Adda di Fagagna F, Falaschi A, Giacca M. Activation of transcription factor NF-B by the Tat protein of human immunodeficiency virus type 1. J. Virol 70 (1996): 4427-4437.
- Demarchi F, Gutierrez MI, Giacca M. 1999. Human immunodeficiency virus type 1 tat protein activates transcription factor NF-B through the cellular interferon-inducible, double-stranded RNA-dependent protein kinase, PKR. J. Virol 73 (1999): 7080 -7086.
- Marzio G, Tyagi M, Gutierrez MI, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl. Acad. Sci 95 (1998): 13519 -13524.
- Treand C, and al. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J 25 (2006): 1690 -1699.
- Vardabasso C, Manganaro L, Lusic M, Marcello A, Giacca M. The histone chaperone protein Nucleosome Assembly Protein-1 (hNAP-1) binds HIV-1 Tat and promotes viral transcription. Retrovirology 5 (2008): 5- 8
- Barboric M, and al. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res 35 (2007): 2003- 2012
- Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 6 (2010): e1001152.
- Daniel A., Donahue, jörn D. Kuhl, Richard D., Sloan, and Mark A. The Viral Protein Tat Can Inhibit the Establishment of HIV-1 Latency. Journal of Virology 86 (2012): 3253-3263
- Siliciano, J.D.; Kajdas, J.; Finzi, D.; Quinn, T.C.; Chadwick, K.; Margolick, J.B.; Kovacs, C.; Gange, S.J.; Siliciano, R.F. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med 9 (2003): 727-728.
- Baxter, A.E.; Niessl, J.; Fromentin, R.; Richard, J.; Porichis, F.; Charlebois, R.; Massanella, M.; Brassard, N.; Alsahafi, N.; Delgado, G.-G.; and al. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe 20 (2016): 368-380.
- Colby, D.J.; Trautmann, L.; Pinyakorn, S.; Leyre, L.; Pagliuzza, A.; Kroon, E.; Rolland, M.; Takata, H.; Buranapraditkun, S.; Intasan, J.; and al. Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat. Med 24 (2018): 923-926.
- Goldstein, G. HIV-1 Tat protein as a potential AIDS vaccine. Nat. Med 2 (1996): 960-964.
- Shi, J.; Manolikakes, G.; Yeh, C.H.; Guerrero, C.A.; Shenvi, R.A.; Shigehisa, H.; Baran, P.S. Scalable synthesis of cortistatin A and related structures. J. Am. Chem. Soc 133 (2011): 8014-8027.
- Mediouni, S.; Chinthalapudi, K.; Ekka, M.K.; Usui, I.; Jablonski, J.A.; Clementz, M.A.; Mousseau, G.; Nowak, J.; Macherla, V.R.; Beverage, J.N. and al. Didehydro-cortistatin A inhibits HIV-1 by specifically binding to the unstructured basic region of Tat. MBio 10 (2019): e02662-18.
- Reiss, P. and al. Speed of progression to AIDS and degree of antibody response to accessory gene products of HIV-1. J. Med. Virol 30 (1990): 163-168.
- Neuveut, C. and Jeang, K.T. Recombinant HIV-1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J. Virol 70 (1996): 5572-5581.
- Ensoli B, Moretti S, Borsetti A, et al. new insights into pathogenesis point to HIV-1 Tat as a key vaccine target.Arch Virol 11 (2021): 2955-2974.
- Paolo Monini, Antonella Tripiciano, Orietta Picconi, Anna Casabianca, Chiara Orland, Sonia Moretti, Vittorio Francavilla, Angela Arancio, Giovanni Paniccia, Massimo Campagna, Stefania Bellino, Marianna Meschiari, Silvia Nozza, Laura Sighinolfi, Alessandra Latini, Antonio Muscatello, Annalisa Saracino, Massimo Di Pietro, Massimo Galli, Aurelio Cafaro, Mauro Magnani Fabrizio Ensoli and Barbara Ensoli. Continued Decay of HIV Proviral DNA Upon Vaccination With HIV-1 Tat of Subjects on Long-Term ART: An 8-Year Follow-Up Study Cecilia Sgadari. Frontiers in Immunology 10 (2019): 1-19.
- AurelioCafaroandBarbaraEnsoli.HIV-1 therapeutic vaccines in clinical development to intensify or replace antiretroviral therapy: the promising results of the Tat vaccine,Expert Review of Vaccines 21 (2022): 1243-1253.





 Impact Factor: * 5.814
Impact Factor: * 5.814 CiteScore: 2.9
CiteScore: 2.9  Acceptance Rate: 11.01%
Acceptance Rate: 11.01%  Time to first decision: 10.4 days
Time to first decision: 10.4 days  Time from article received to acceptance: 2-3 weeks
Time from article received to acceptance: 2-3 weeks 